Structural Analysis of Escherichia coli OpgG, a Protein Required for the Biosynthesis of Osmoregulated Periplasmic Glucans.
Hanoulle, X., Rollet, E., Clantin, B., Landrieu, I., Odberg-Ferragut, C., Lippens, G., Bohin, J.P., Villeret, V.(2004) J Mol Biol 342: 195-205
- PubMed: 15313617
- DOI: https://doi.org/10.1016/j.jmb.2004.07.004
- Primary Citation of Related Structures:
1TXK - PubMed Abstract:
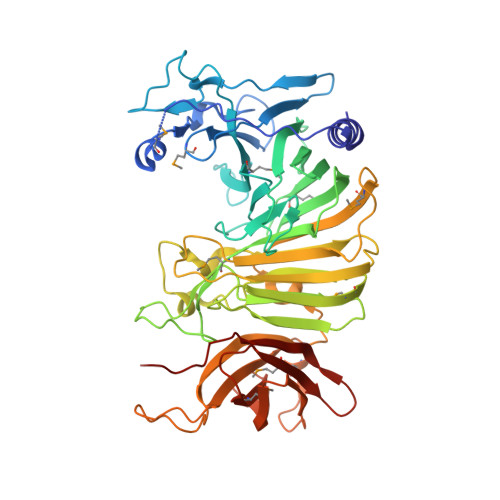
Osmoregulated periplasmic glucans (OPGs) G protein (OpgG) is required for OPGs biosynthesis. OPGs from Escherichia coli are branched glucans, with a backbone of beta-1,2 glucose units and with branches attached by beta-1,6 linkages. In Proteobacteria, OPGs are involved in osmoprotection, biofilm formation, virulence and resistance to antibiotics. Despite their important biological implications, enzymes synthesizing OPGs are poorly characterized. Here, we report the 2.5 A crystal structure of OpgG from E.coli. The structure was solved using a selenemethionine derivative of OpgG and the multiple anomalous diffraction method (MAD). The protein is composed of two beta-sandwich domains connected by one turn of 3(10) helix. The N-terminal domain (residues 22-388) displays a 25-stranded beta-sandwich fold found in several carbohydrate-related proteins. It exhibits a large cleft comprising many aromatic and acidic residues. This putative binding site shares some similarities with enzymes such as galactose mutarotase and glucodextranase, suggesting a potential catalytic role for this domain in OPG synthesis. On the other hand, the C-terminal domain (residues 401-512) has a seven-stranded immunoglobulin-like beta-sandwich fold, found in many proteins where it is mainly implicated in interactions with other molecules. The structural data suggest that OpgG is an OPG branching enzyme in which the catalytic activity is located in the large N-terminal domain and controlled via the smaller C-terminal domain.
Organizational Affiliation:
UMR 8525 CNRS, Institut de Biologie de Lille, Université de Lille II, 1 rue du Professeur Calmette, BP447, 59021, France. xavier.hanoulle@ibl.fr