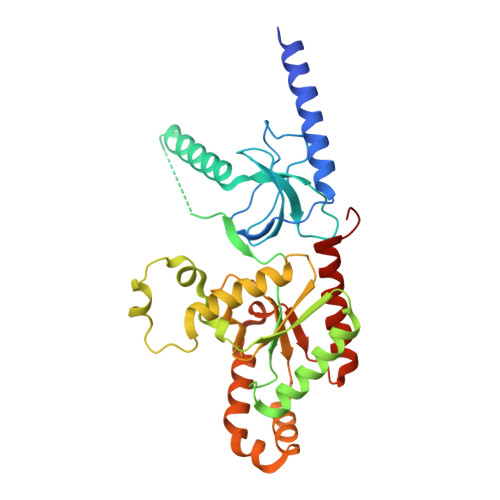
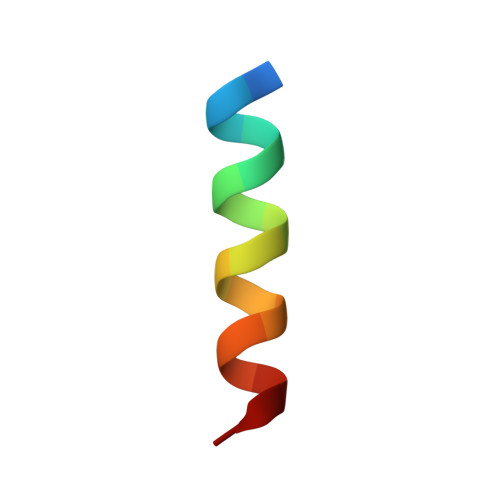Structural Analysis of Voltage-Dependent Calcium Channel Beta Subunit Functional Core and Its Complex with the Alpha1 Interaction Domain
Opatowsky, Y., Chen, C.-C., Campbell, K.P., Hirsch, J.A.(2004) Neuron 42: 387-399
- PubMed: 15134636
- DOI: https://doi.org/10.1016/s0896-6273(04)00250-8
- Primary Citation of Related Structures:
1T3L, 1T3S - PubMed Abstract:
Voltage-dependent calcium channels (VDCC) are multiprotein assemblies that regulate the entry of extracellular calcium into electrically excitable cells and serve as signal transduction centers. The alpha1 subunit forms the membrane pore while the intracellular beta subunit is responsible for trafficking of the channel to the plasma membrane and modulation of its electrophysiological properties. Crystallographic analyses of a beta subunit functional core alone and in complex with a alpha1 interaction domain (AID) peptide, the primary binding site of beta to the alpha1 subunit, reveal that beta represents a novel member of the MAGUK protein family. The findings illustrate how the guanylate kinase fold has been fashioned into a protein-protein interaction module by alteration of one of its substrate sites. Combined results indicate that the AID peptide undergoes a helical transition in binding to beta. We outline the mechanistic implications for understanding the beta subunit's broad regulatory role of the VDCC, particularly via the AID.
Organizational Affiliation:
Department of Biochemistry, Faculty of Life Sciences, Tel Aviv University, Ramat Aviv 69978, Israel.