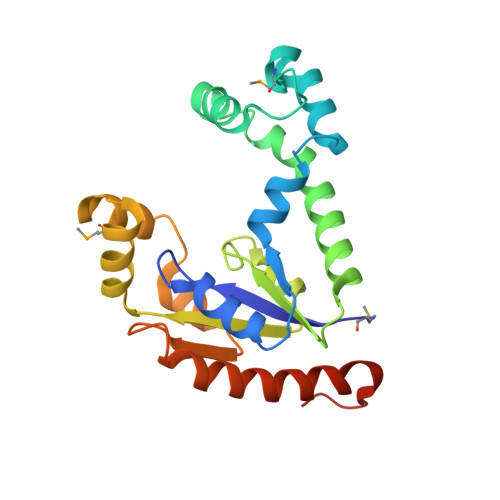
X-ray Structure of Dephospho-CoA Kinase from E. coli Norteast Structural Genomics Consortium Target ER57
Kuzin, A.P., Chen, Y., Forouhar, F., Edstrom, W., Benach, J., Vorobiev, S., Acton, T., Shastry, R., Ma, L.-C., Xia, R., Montelione, G., Tong, L., Hunt, J.To be published.