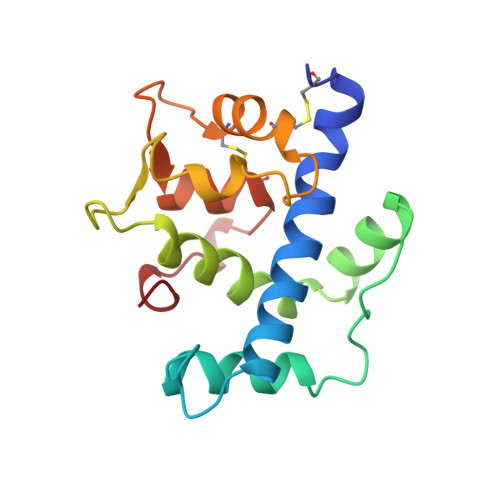
Structure of a novel extracellular Ca(2+)-binding module in BM-40.
Hohenester, E., Maurer, P., Hohenadl, C., Timpl, R., Jansonius, J.N., Engel, J.(1996) Nat Struct Biol 3: 67-73
- PubMed: 8548457
- DOI: https://doi.org/10.1038/nsb0196-67
- Primary Citation of Related Structures:
1SRA - PubMed Abstract:
The EF-hand is a highly conserved Ca(2+)-binding motif found in many cytosolic Ca(2+)-modulated proteins. Here we report the crystal structure at 2.0 A resolution of the carboxy-terminal domain of human BM-40 (SPARC, osteonectin), an extracellular matrix protein containing an EF-hand pair. The two EF-hands interact canonically but their detailed structures are unusual. In the first EF-hand a one-residue insertion is accommodated by a cis-peptide bond and by substituting a carboxylate by a peptide carbonyl as a Ca2+ ligand. The second EF-hand is stabilized by a disulphide bond. The EF-hand pair interacts tightly with an amphiphilic amino-terminal helix, reminiscent of target peptide binding by calmodulin. The present structure defines a novel protein module occurring in several other extracellular proteins.
Organizational Affiliation:
Department of Structural Biology, University of Basel, Switzerland.