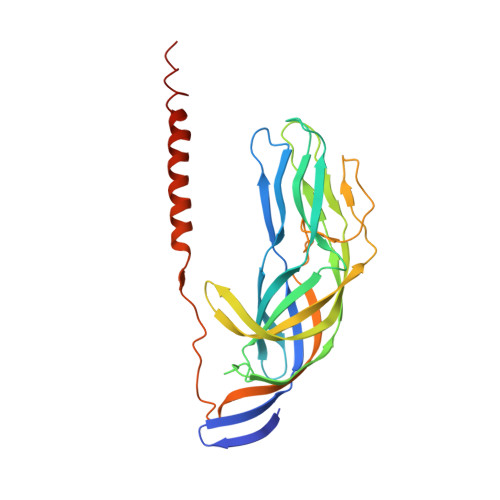
Structural rearrangements in the membrane penetration protein of a non-enveloped virus.
Dormitzer, P.R., Nason, E.B., Prasad, B.V., Harrison, S.C.(2004) Nature 430: 1053-1058
- PubMed: 15329727
- DOI: https://doi.org/10.1038/nature02836
- Primary Citation of Related Structures:
1SLQ - PubMed Abstract:
Non-enveloped virus particles (those that lack a lipid-bilayer membrane) must breach the membrane of a target host cell to gain access to its cytoplasm. So far, the molecular mechanism of this membrane penetration step has resisted structural analysis. The spike protein VP4 is a principal component in the entry apparatus of rotavirus, a non-enveloped virus that causes gastroenteritis and kills 440,000 children each year. Trypsin cleavage of VP4 primes the virus for entry by triggering a rearrangement that rigidifies the VP4 spikes. We have determined the crystal structure, at 3.2 A resolution, of the main part of VP4 that projects from the virion. The crystal structure reveals a coiled-coil stabilized trimer. Comparison of this structure with the two-fold clustered VP4 spikes in a approximately 12 A resolution image reconstruction from electron cryomicroscopy of trypsin-primed virions shows that VP4 also undergoes a second rearrangement, in which the oligomer reorganizes and each subunit folds back on itself, translocating a potential membrane-interaction peptide from one end of the spike to the other. This rearrangement resembles the conformational transitions of membrane fusion proteins of enveloped viruses.
Organizational Affiliation:
Department of Pediatrics, Harvard Medical School, and the Laboratory of Molecular Medicine, Children's Hospital, Boston, Massachusetts 02115, USA. dormitze@crystal.harvard.edu