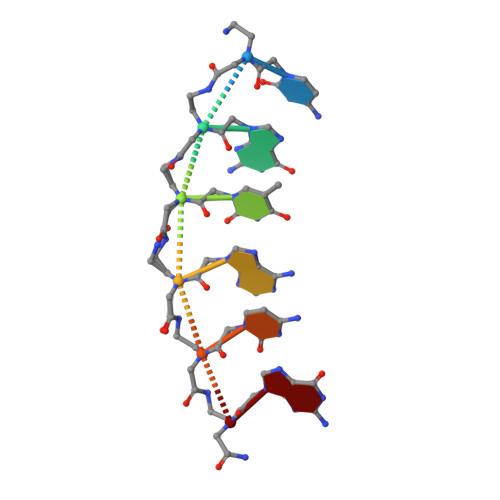
Crystal structure of a peptide nucleic acid (PNA) duplex at 1.7 A resolution.
Rasmussen, H., Kastrup, J.S., Nielsen, J.N., Nielsen, J.M., Nielsen, P.E.(1997) Nat Struct Biol 4: 98-101
- PubMed: 9033585
- DOI: https://doi.org/10.1038/nsb0297-98
- Primary Citation of Related Structures:
1PUP - PubMed Abstract:
The crystal structure of a PNA duplex reveals both a right- and a left-handed helix in the unit cell. The helices are wide (28A), large pitched (18bp) with the base pairs perpendicular to the helix axis, thereby demonstrating that PNA besides adapting to oligonucleotide partners also has a unique structure by itself.