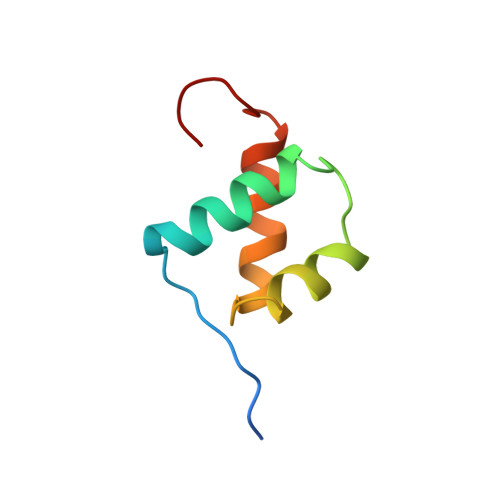
Solution structure of the Oct-1 POU homeodomain determined by NMR and restrained molecular dynamics.
Cox, M., van Tilborg, P.J., de Laat, W., Boelens, R., van Leeuwen, H.C., van der Vliet, P.C., Kaptein, R.(1995) J Biomol NMR 6: 23-32
- PubMed: 7663141
- DOI: https://doi.org/10.1007/BF00417488
- Primary Citation of Related Structures:
1POG - PubMed Abstract:
The POU homeodomain (POUhd), a divergent member of the well-studied class of homeodomain proteins, is the C-terminal part of the bipartite POU domain, the conserved DNA-binding domain of the POU proteins. In this paper we present the solution structure of POUhd of the human Oct-1 transcription factor. This fragment was overexpressed in Escherichia coli and studied by two- and three-dimensional homo- and heteronuclear NMR techniques, resulting in virtually complete 1H and 15N resonance assignments for residues 2-60. Using distance and dihedral constraints derived from the NMR data, 50 distance geometry structures were calculated, which were refined by means of restrained molecular dynamics. From this set a total of 31 refined structures were selected, having low constraint energy and few constraint violations. The ensemble of 31 structures displays a root-mean-square deviation of the coordinates of 0.59 A with respect to the average structure, calculated over the backbone atoms of residues 6 to 54. The fold of POUhd is very similar to that of the canonical homeodomains. Interestingly, the recognition helix of the free POUhd ends at residue 53, while in the cocrystal structure of the intact POU domain with the DNA octamer motif [Klemm, J.D., Rould, M.A., Aurora, R., Herr, W. and Pabo, C.O. (1994) Cell, 77, 21-32] this helix in the POUhd subdomain is extended as far as residue 60.
Organizational Affiliation:
Bijvoet Center for Biomolecular Research, Utrecht University, The Netherlands.