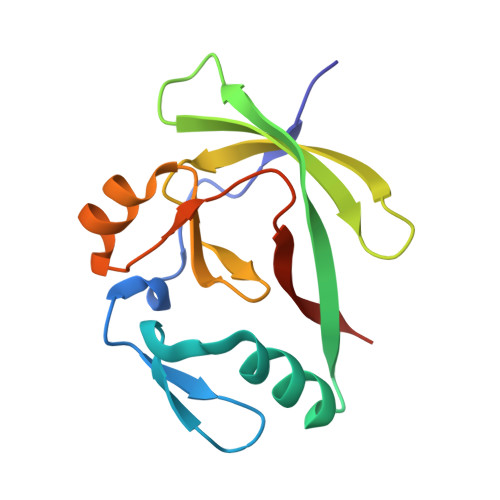
The structure of the AXH domain of spinocerebellar ataxin-1.
Chen, Y.W., Allen, M.D., Veprintsev, D.B., Lowe, J., Bycroft, M.(2004) J Biol Chem 279: 3758-3765
- PubMed: 14583607
- DOI: https://doi.org/10.1074/jbc.M309817200
- Primary Citation of Related Structures:
1OA8 - PubMed Abstract:
Spinocerebellar ataxia type 1 is a late-onset neurodegenerative disease caused by the expansion of a CAG triplet repeat in the SCA1 gene. This results in the lengthening of a polyglutamine tract in the gene product ataxin-1. This produces a toxic gain of function that results in specific neuronal death. A region in ataxin-1, the AXH domain, exhibits significant sequence similarity to the transcription factor HBP1. This region of the protein has been implicated in RNA binding and self-association. We have determined the crystal structure of the AXH domain of ataxin-1. The AXH domain is dimeric and contains an OB-fold, a structural motif found in many oligonucleotide-binding proteins, supporting its proposed role in RNA binding. By structure comparison with other proteins that contain an OB-fold, a putative RNA-binding site has been identified. We also identified a cluster of charged surface residues that are well conserved among AXH domains. These residues may constitute a second ligand-binding surface, suggesting that all AXH domains interact with a common yet unidentified partner.
Organizational Affiliation:
Centre for Protein Engineering, Medical Research Council Centre, Cambridge, UK.