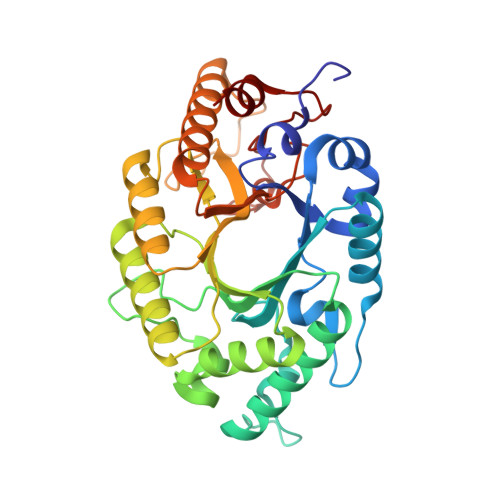
The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus
Solomon, V., Teplitsky, A., Golan, G., Gilboa, R., Reiland, V., Shulami, S., Moryles, S., Zolotnitsky, G., Shoham, Y., Shoham, G.To be published.