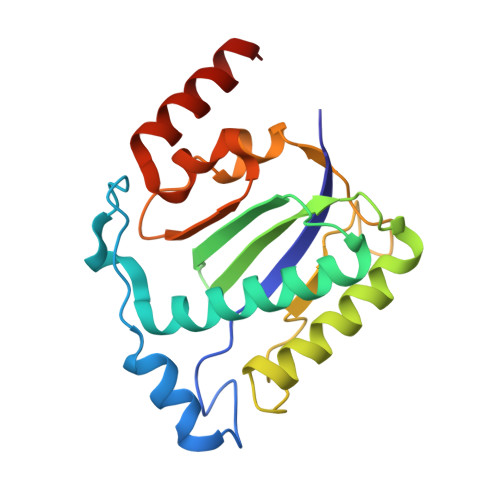
Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep.
Hickman, A.B., Ronning, D.R., Kotin, R.M., Dyda, F.(2002) Mol Cell 10: 327-337
- PubMed: 12191478
- DOI: https://doi.org/10.1016/s1097-2765(02)00592-0
- Primary Citation of Related Structures:
1M55 - PubMed Abstract:
Adeno-associated virus (AAV), unique among animal viruses in its ability to integrate into a specific chromosomal location, is a promising vector for human gene therapy. AAV Replication (Rep) protein is essential for viral replication and integration, and its amino terminal domain possesses site-specific DNA binding and endonuclease activities required for replication initiation and integration. This domain displays a novel endonuclease fold and demonstrates an unexpected structural relationship to other viral origin binding proteins such as the papillomavirus E1 protein and the SV40 T antigen. The active site, located at the bottom of a positively charged cleft, is formed by the spatial convergence of a divalent metal ion and two conserved sequence motifs that define the rolling circle replication superfamily.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA. ahickman@helix.nih.gov