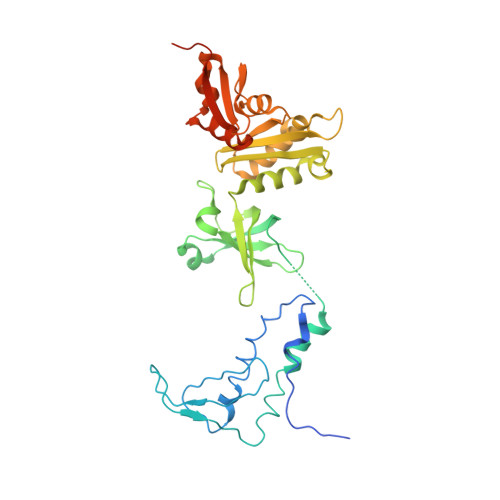
Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 A resolution.
Gopal, B., Haire, L.F., Gamblin, S.J., Dodson, E.J., Lane, A.N., Papavinasasundaram, K.G., Colston, M.J., Dodson, G.(2001) J Mol Biol 314: 1087-1095
- PubMed: 11743725
- DOI: https://doi.org/10.1006/jmbi.2000.5144
- Primary Citation of Related Structures:
1K0R - PubMed Abstract:
Mycobacterium tuberculosis is the cause of tuberculosis in humans, a disease that affects over a one-third of the world's population. This slow-growing pathogen has only one ribosomal RNA operon, thus making its transcriptional apparatus a fundamentally interesting target for drug discovery. NusA binds to RNA polymerase and modulates several of the ribosomal RNA transcriptional processes. Here, we report the crystal structure of NusA, and reveal that the molecule consists of four domains. They are organised as two distinct entities. The N-terminal domain (residues 1 to 99) that resembles the B chain of the Rad50cd ATP binding cassette-ATPase (ABC-ATPase) and a C-terminal module (residues 108 to 329) consisting of a ribosomal S1 protein domain followed by two K homology domains. The S1 and KH domains are tightly integrated together to form an extensive RNA-binding structure, but are flexibly tethered to the N-terminal domain. The molecule's surfaces and architecture provide insights into RNA and polymerase interactions and the mechanism of pause site discrimination. They also allow us to rationalize certain termination-defective and cold shock-sensitive mutations in the nusA gene that have been studied in Escherichia coli.
Organizational Affiliation:
Division of Protein Structure, National Institute for Medical Research, Mill Hill, London, UK.