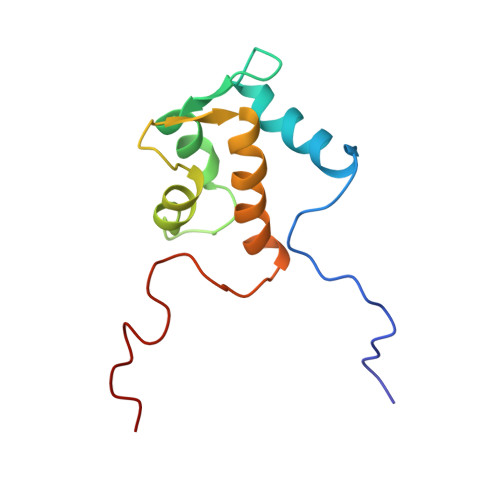
Solution structure of the Eps15 homology domain of a human POB1 (partner of RalBP1).
Koshiba, S., Kigawa, T., Iwahara, J., Kikuchi, A., Yokoyama, S.(1999) FEBS Lett 442: 138-142
- PubMed: 9928989
- DOI: https://doi.org/10.1016/s0014-5793(98)01644-5
- Primary Citation of Related Structures:
1IQ3 - PubMed Abstract:
The solution structure of the Eps15 homology (EH) domain of a human POB1 (partner of RaIBP1) has been determined by uniform 13C/15N labeling and heteronuclear multidimensional nuclear magnetic resonance spectroscopy. The POB1 EH domain consists of two EF-hand structures, and the second one binds a calcium ion. In the calcium-bound state, the orientation of the fourth alpha-helix relative to the other helices of the POB1 EH domain is slightly different from that of calbindin, and much more different from those of calmodulin and troponin C, on the basis of their atomic coordinates.
Organizational Affiliation:
Cellular Signaling Laboratory, The Institute of Physical and Chemical Research (RIKEN), Wako, Saitama, Japan.