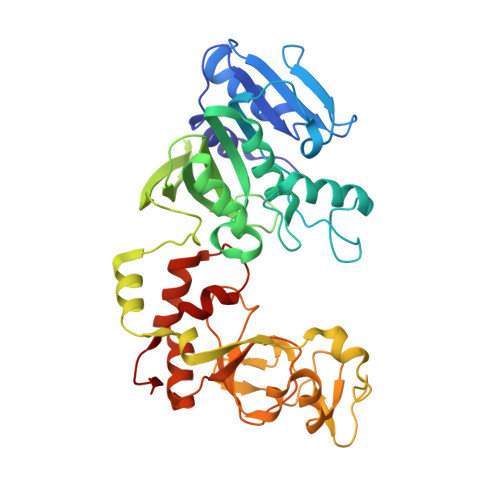
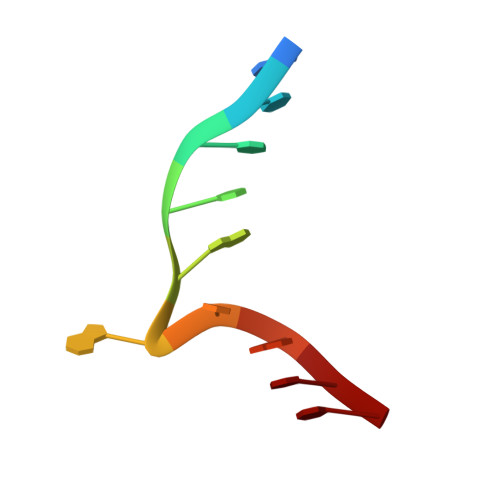
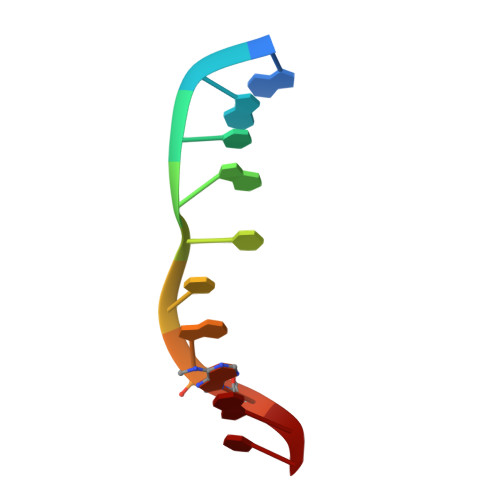
Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog.
Goedecke, K., Pignot, M., Goody, R.S., Scheidig, A.J., Weinhold, E.(2001) Nat Struct Biol 8: 121-125
- PubMed: 11175899
- DOI: https://doi.org/10.1038/84104
- Primary Citation of Related Structures:
1G38 - PubMed Abstract:
The 2.0 A crystal structure of the N6-adenine DNA methyltransferase M.TaqI in complex with specific DNA and a nonreactive cofactor analog reveals a previously unrecognized stabilization of the extrahelical target base. To catalyze the transfer of the methyl group from the cofactor S-adenosyl-l-methionine to the 6-amino group of adenine within the double-stranded DNA sequence 5'-TCGA-3', the target nucleoside is rotated out of the DNA helix. Stabilization of the extrahelical conformation is achieved by DNA compression perpendicular to the DNA helix axis at the target base pair position and relocation of the partner base thymine in an interstrand pi-stacked position, where it would sterically overlap with an innerhelical target adenine. The extrahelical target adenine is specifically recognized in the active site, and the 6-amino group of adenine donates two hydrogen bonds to Asn 105 and Pro 106, which both belong to the conserved catalytic motif IV of N6-adenine DNA methyltransferases. These hydrogen bonds appear to increase the partial negative charge of the N6 atom of adenine and activate it for direct nucleophilic attack on the methyl group of the cofactor.
Organizational Affiliation:
Max-Planck-Institut für molekulare Physiologie, Abteilung Physikalische Biochemie, Otto-Hahn-Str. 11, D-44227 Dortmund, Germany.