Interruption of the internal water chain of cytochrome f impairs photosynthetic function.
Sainz, G., Carrell, C.J., Ponamarev, M.V., Soriano, G.M., Cramer, W.A., Smith, J.L.(2000) Biochemistry 39: 9164-9173
- PubMed: 10924110
- DOI: https://doi.org/10.1021/bi0004596
- Primary Citation of Related Structures:
1E2V, 1E2W, 1E2Z, 1EWH - PubMed Abstract:
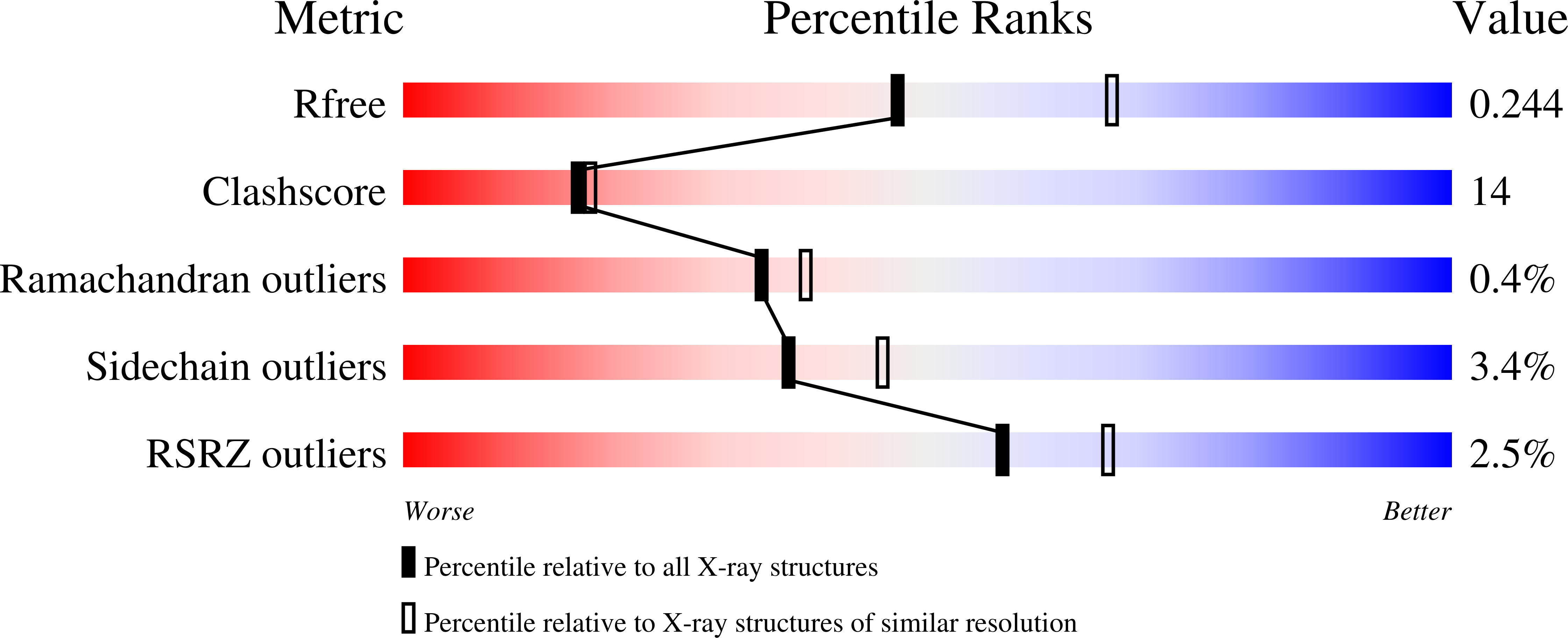
The structure of cytochrome f includes an internal chain of five water molecules and six hydrogen-bonding side chains, which are conserved throughout the phylogenetic range of photosynthetic organisms from higher plants, algae, and cyanobacteria. The in vivo electron transfer capability of Chlamydomonas reinhardtii cytochrome f was impaired in site-directed mutants of the conserved Asn and Gln residues that form hydrogen bonds with water molecules of the internal chain [Ponamarev, M. V., and Cramer, W. A. (1998) Biochemistry 37, 17199-17208]. The 251-residue extrinsic functional domain of C. reinhardtii cytochrome f was expressed in Escherichia coli without the 35 C-terminal residues of the intact cytochrome that contain the membrane anchor. Crystal structures were determined for the wild type and three "water chain" mutants (N168F, Q158L, and N153Q) having impaired photosynthetic and electron transfer function. The mutant cytochromes were produced, folded, and assembled heme at levels identical to that of the wild type in the E. coli expression system. N168F, which had a non-photosynthetic phenotype and was thus most affected by mutational substitution, also had the greatest structural perturbation with two water molecules (W4 and W5) displaced from the internal chain. Q158L, the photosynthetic mutant with the largest impairment of in vivo electron transfer, had a more weakly bound water at one position (W1). N153Q, a less impaired photosynthetic mutant, had an internal water chain with positions and hydrogen bonds identical to those of the wild type. The structure data imply that the waters of the internal chain, in addition to the surrounding protein, have a significant role in cytochrome f function.
Organizational Affiliation:
Department of Biological Sciences, Purdue University, West Lafayette, Indiana 47907-1392, USA.