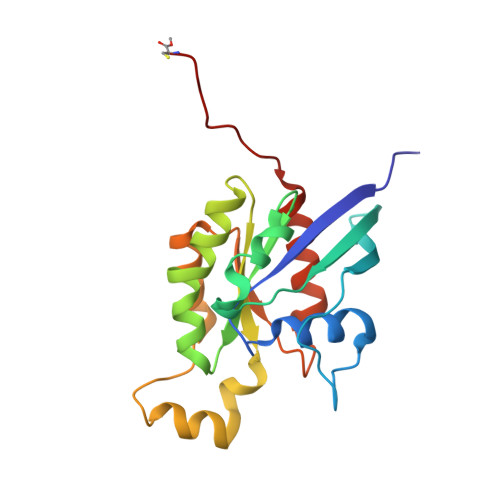
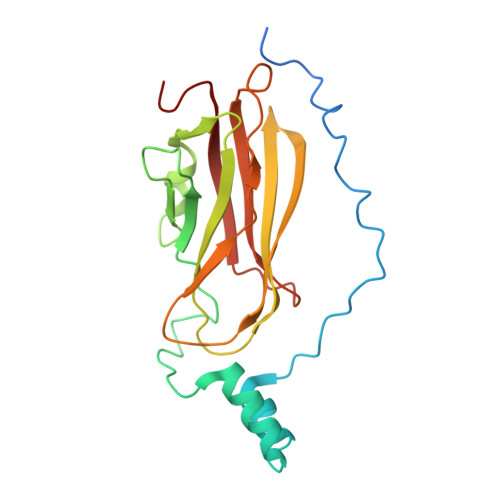
Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI.
Hoffman, G.R., Nassar, N., Cerione, R.A.(2000) Cell 100: 345-356
- PubMed: 10676816
- DOI: https://doi.org/10.1016/s0092-8674(00)80670-4
- Primary Citation of Related Structures:
1DOA - PubMed Abstract:
The RhoGDI proteins serve as key multifunctional regulators of Rho family GTP-binding proteins. The 2.6 A X-ray crystallographic structure of the Cdc42/RhoGDI complex reveals two important sites of interaction between GDI and Cdc42. First, the amino-terminal regulatory arm of the GDI binds to the switch I and II domains of Cdc42 leading to the inhibition of both GDP dissociation and GTP hydrolysis. Second, the geranylgeranyl moiety of Cdc42 inserts into a hydrophobic pocket within the immunoglobulin-like domain of the GDI molecule leading to membrane release. The structural data demonstrate how GDIs serve as negative regulators of small GTP-binding proteins and how the isoprenoid moiety is utilized in this critical regulatory interaction.
Organizational Affiliation:
Department of Molecular Medicine, Veterinary Medical Center, Cornell University, Ithaca, New York 14853, USA.