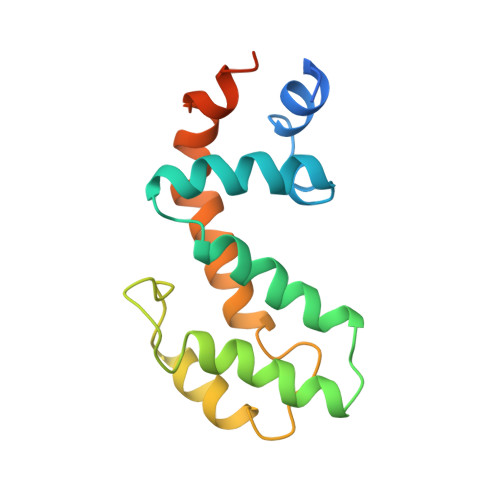
Solution structure of human GAIP (Galpha interacting protein): a regulator of G protein signaling.
de Alba, E., De Vries, L., Farquhar, M.G., Tjandra, N.(1999) J Mol Biol 291: 927-939
- PubMed: 10452897
- DOI: https://doi.org/10.1006/jmbi.1999.2989
- Primary Citation of Related Structures:
1CMZ - PubMed Abstract:
The solution structure of the human protein GAIP (Galpha interacting protein), a regulator of G protein signaling, has been determined by NMR techniques. Dipolar couplings of the oriented protein in two different liquid crystal media have been used in the structure calculation. The solution structure of GAIP is compared to the crystal structure of an homologous protein from rat (RGS4) complexed to the alpha-subunit of a G protein. Some of RGS4 residues involved in the Galpha-RGS binding interface have similar orientations in GAIP (free form), indicating that upon binding these residues do not suffer conformational rearrangements, and therefore, their role does not seem to be restricted to Galpha interaction but also to RGS folding and stability. We suggest that other structural differences between the two proteins may be related to the process of binding as well as to a distinct efficiency in their respective GTPase activating function.
Organizational Affiliation:
Laboratory of Biophysical Chemistry, Building 3 National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD, 20892-0380, USA.