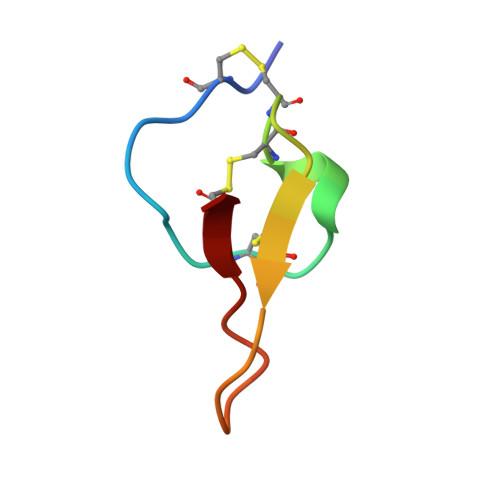
A new fold in the scorpion toxin family, associated with an activity on a ryanodine-sensitive calcium channel.
Mosbah, A., Kharrat, R., Fajloun, Z., Renisio, J.G., Blanc, E., Sabatier, J.M., El Ayeb, M., Darbon, H.(2000) Proteins 40: 436-442
- PubMed: 10861934
- DOI: https://doi.org/10.1002/1097-0134(20000815)40:3<436::aid-prot90>3.0.co;2-9
- Primary Citation of Related Structures:
1C6W - PubMed Abstract:
We determined the structure in solution by (1)H two-dimensional NMR of Maurocalcine from the venom of Scorpio maurus. This toxin has been demonstrated to be a potent effector of ryanodyne-sensitive calcium channel from skeletal muscles. This is the first description of a scorpion toxin which folds following the Inhibitor Cystine Knot fold (ICK) already described for numerous toxic and inhibitory peptides, as well as for various protease inhibitors. Its three dimensional structure consists of a compact disulfide-bonded core from which emerge loops and the N-terminus. A double-stranded antiparallel beta-sheet comprises residues 20-23 and 30-33. A third extended strand (residues 9-11) is perpendicular to the beta-sheet. Maurocalcine structure mimics the activating segment of the dihydropyridine receptor II-III loop and is therefore potentially useful for dihydropyridine receptor/ryanodine receptor interaction studies. Proteins 2000;40:436-442.
Organizational Affiliation:
AFMB, Marseille, France.