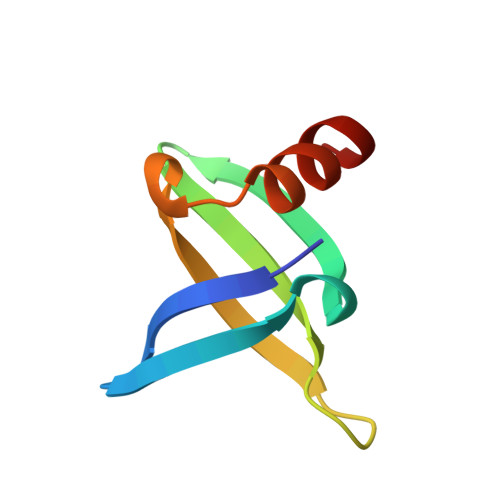
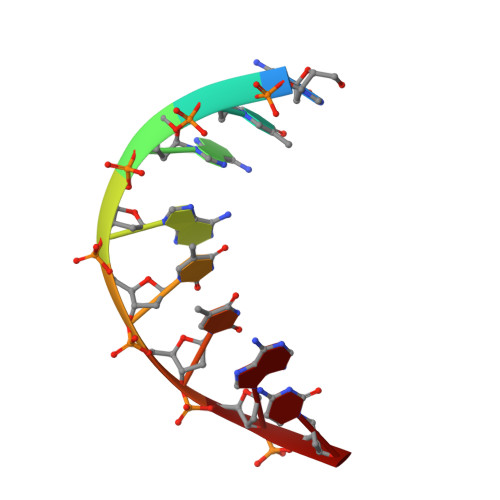
The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA.
Gao, Y.G., Su, S.Y., Robinson, H., Padmanabhan, S., Lim, L., McCrary, B.S., Edmondson, S.P., Shriver, J.W., Wang, A.H.(1998) Nat Struct Biol 5: 782-786
- PubMed: 9731772
- DOI: https://doi.org/10.1038/1822
- Primary Citation of Related Structures:
1BF4, 1BNZ - PubMed Abstract:
Sso7d and Sac7d are two small (approximately 7,000 Mr), but abundant, chromosomal proteins from the hyperthermophilic archaeabacteria Sulfolobus solfataricus and S. acidocaldarius respectively. These proteins have high thermal, acid and chemical stability. They bind DNA without marked sequence preference and increase the Tm of DNA by approximately 40 degrees C. Sso7d in complex with GTAATTAC and GCGT(iU)CGC + GCGAACGC was crystallized in different crystal lattices and the crystal structures were solved at high resolution. Sso7d binds in the minor groove of DNA and causes a single-step sharp kink in DNA (approximately 60 degrees) by the intercalation of the hydrophobic side chains of Val 26 and Met 29. The intercalation sites are different in the two complexes. Observations of this novel DNA binding mode in three independent crystal lattices indicate that it is not a function of crystal packing.
Organizational Affiliation:
Department of Cell and Structural Biology, University of Illinois at Urbana-Champaign, Urbana 61801, USA.