New Features
Integrating PDB Data with NIH Common Fund Data Resources
07/21
RCSB PDB has integrated three-dimensional structural information with 3 new resources that fall under the umbrella of the NIH Common Fund Data Resources. Ultimately, this integration is expected to enable investigation of novel biological questions, and promote a more complete understanding of human health and disease.
Relevant PDB structures are now linked to individual data in PHAROS, GTEx, and IMPC.
- PHAROS is part of the Integrating the Druggable Genome (IDG) project and presents integrated data for 3 of the most important drug target protein families: GPCRs, ion channels and kinases. Some of the data types it covers include: disease, phenotypic and ontologies such as the Drug Target Ontology.
- GTEx is a public resource for the study of tissue-specific gene expression and regulation.
- IMPC covers data related to mouse phenotypes, as part of an international effort by 19 research institutions to identify the function of every protein-coding gene in the mouse genome.
Data are accessible from individual Structure Summary pages and Advanced Search.
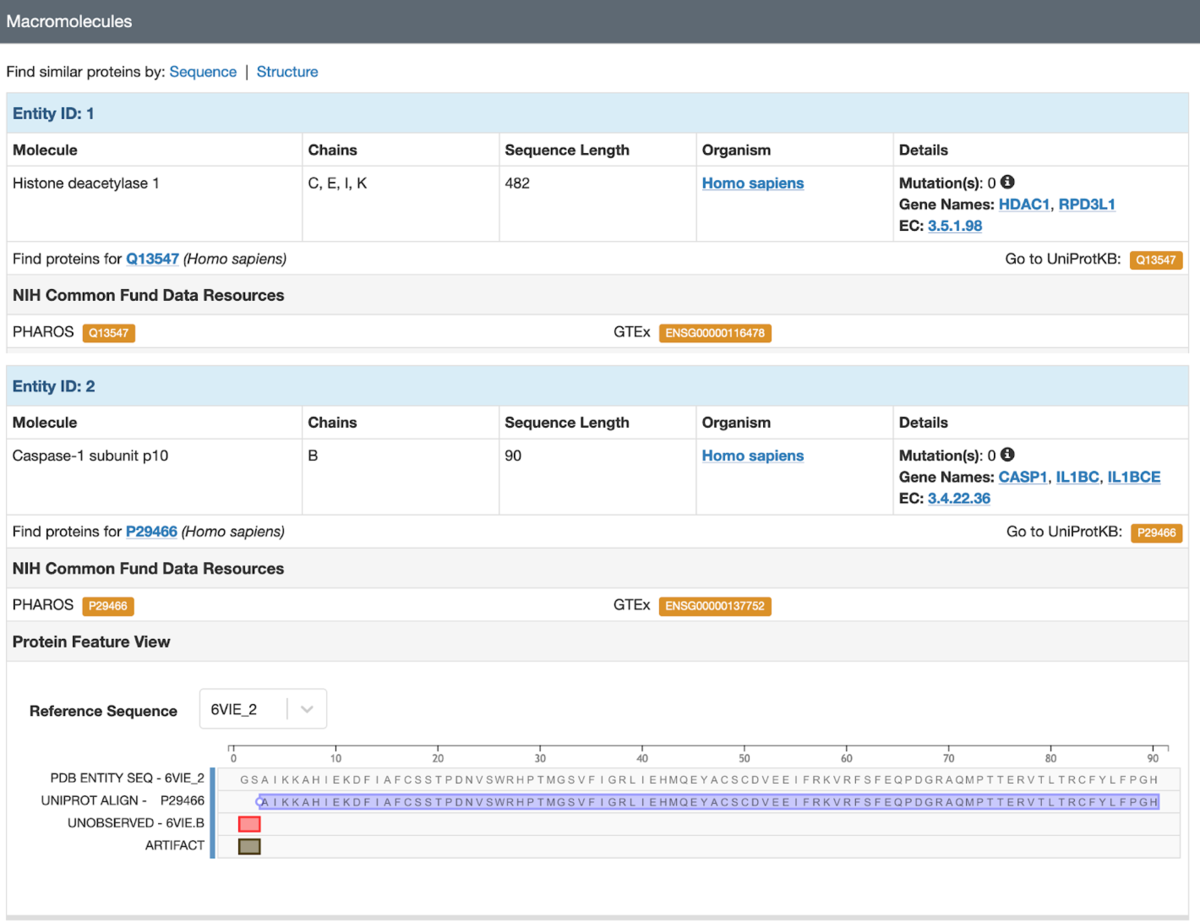 The Structure Summary page for PDB structure 6vie (Structure of caspase-1 in complex with gasdermin D) links to corresponding pages at PHAROS and GTEx for Entities 1 and 2, and to IMPC for Entity 3.
The Structure Summary page for PDB structure 6vie (Structure of caspase-1 in complex with gasdermin D) links to corresponding pages at PHAROS and GTEx for Entities 1 and 2, and to IMPC for Entity 3. 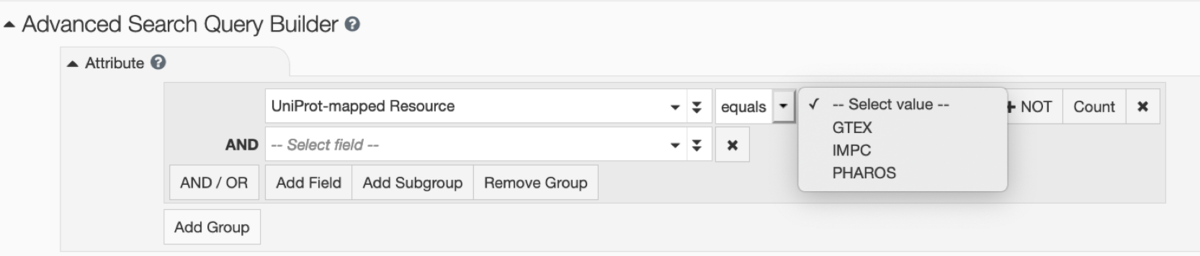 Searching PDB entries that have a mapping to these 3 NIH Common Fund data resources is possible in the RCSB.org Advanced Search User Interface by selecting Attribute: Linked External Resources and UniProt-mapped Resource.
Searching PDB entries that have a mapping to these 3 NIH Common Fund data resources is possible in the RCSB.org Advanced Search User Interface by selecting Attribute: Linked External Resources and UniProt-mapped Resource.This work was supported by an NIH Common Fund Data Use Award intended for Enhancing Utility and Usage of Common Fund Data Sets.












