Medicinal Au(I) compounds targeting urease as prospective antimicrobial agents: unveiling the structural basis for enzyme inhibition.
Mazzei, L., Massai, L., Cianci, M., Messori, L., Ciurli, S.(2021) Dalton Trans 50: 14444-14452
- PubMed: 34585201
- DOI: https://doi.org/10.1039/d1dt02488d
- Primary Citation of Related Structures:
7P7N, 7P7O - PubMed Abstract:
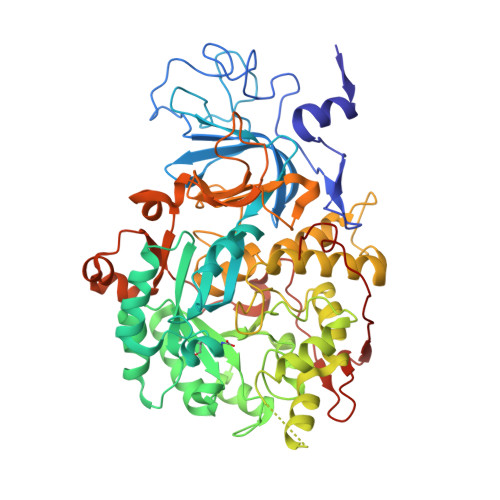
A few gold compounds were recently found to show antimicrobial properties in vitro , holding great promise for the discovery of new drugs to overcome antibiotic resistance. Here, the inhibition of the bacterial virulence factor urease by four Au(I)-compounds, namely Au(PEt 3 )Cl, Au(PEt 3 )Br, Au(PEt 3 )I and [Au(PEt 3 ) 2 ]Cl, obtained from the antiarthritic Au(I)-drug Auranofin and earlier reported to act as antimicrobials, is investigated. The three monophosphino Au(I) complexes showed IC 50 values in the 30-100 nM range, while the diphosphino Au(I) complex, though being less active, still showed a IC 50 value of 7 μM. The structural basis for this inhibition was provided by solving the crystal structures of urease co-crystallized with Au(PEt 3 )I and [Au(PEt 3 ) 2 ]Cl: at least two Au(I) ions bind the enzyme in a flap domain involved in the catalysis, thus obliterating enzyme activity. Peculiar changes observed in the two structures reveal implications for the mechanism of soft metal binding and enzyme inactivation.
Organizational Affiliation:
Laboratory of Bioinorganic Chemistry, Department of Pharmacy and Biotechnology (FaBiT), University of Bologna, Via Giuseppe Fanin 40, I-40127 Bologna, Italy. luca.mazzei2@unibo.it.