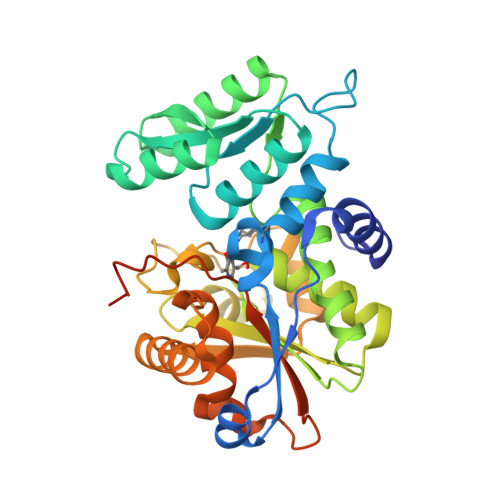
Tyrosine 121 moves revealing a ligandable pocket that couples catalysis to ATP-binding in serine racemase.
Koulouris, C.R., Gardiner, S.E., Harris, T.K., Elvers, K.T., Mark Roe, S., Gillespie, J.A., Ward, S.E., Grubisha, O., Nicholls, R.A., Atack, J.R., Bax, B.D.(2022) Commun Biol 5: 346-346
- PubMed: 35410329
- DOI: https://doi.org/10.1038/s42003-022-03264-5
- Primary Citation of Related Structures:
6ZSP, 6ZUJ, 7NBC, 7NBD, 7NBF, 7NBG, 7NBH - PubMed Abstract:
Human serine racemase (hSR) catalyses racemisation of L-serine to D-serine, the latter of which is a co-agonist of the NMDA subtype of glutamate receptors that are important in synaptic plasticity, learning and memory. In a 'closed' hSR structure containing the allosteric activator ATP, the inhibitor malonate is enclosed between the large and small domains while ATP is distal to the active site, residing at the dimer interface with the Tyr121 hydroxyl group contacting the α-phosphate of ATP. In contrast, in 'open' hSR structures, Tyr121 sits in the core of the small domain with its hydroxyl contacting the key catalytic residue Ser84. The ability to regulate SR activity by flipping Tyr121 from the core of the small domain to the dimer interface appears to have evolved in animals with a CNS. Multiple X-ray crystallographic enzyme-fragment structures show Tyr121 flipped out of its pocket in the core of the small domain. Data suggest that this ligandable pocket could be targeted by molecules that inhibit enzyme activity.
Organizational Affiliation:
Sussex Drug Discovery Centre, University of Sussex, Brighton, BN1 9QG, UK.