Structural and functional insights into iron acquisition from lactoferrin and transferrin in Gram-negative bacterial pathogens.
Chan, C., Ng, D., Fraser, M.E., Schryvers, A.B.(2022) Biometals
- PubMed: 36418809
- DOI: https://doi.org/10.1007/s10534-022-00466-6
- Primary Citation of Related Structures:
7LI0, 7LI1 - PubMed Abstract:
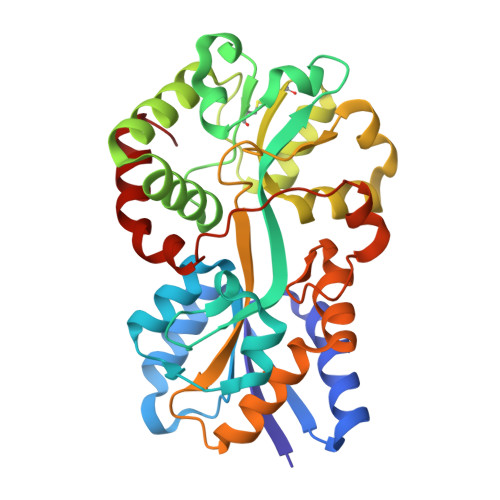
Iron is an essential element for various lifeforms but is largely insoluble due to the oxygenation of Earth's atmosphere and oceans during the Proterozoic era. Metazoans evolved iron transport glycoproteins, like transferrin (Tf) and lactoferrin (Lf), to keep iron in a non-toxic, usable form, while maintaining a low free iron concentration in the body that is unable to sustain bacterial growth. To survive on the mucosal surfaces of the human respiratory tract where it exclusively resides, the Gram-negative bacterial pathogen Moraxella catarrhalis utilizes surface receptors for acquiring iron directly from human Tf and Lf. The receptors are comprised of a surface lipoprotein to capture iron-loaded Tf or Lf and deliver it to a TonB-dependent transporter (TBDT) for removal of iron and transport across the outer membrane. The subsequent transport of iron into the cell is normally mediated by a periplasmic iron-binding protein and inner membrane transport complex, which has yet to be determined for Moraxella catarrhalis. We identified two potential periplasm to cytoplasm transport systems and performed structural and functional studies with the periplasmic binding proteins (FbpA and AfeA) to evaluate their role. Growth studies with strains deleted in the fbpA or afeA gene demonstrated that FbpA, but not AfeA, was required for growth on human Tf or Lf. The crystal structure of FbpA with bound iron in the open conformation was obtained, identifying three tyrosine ligands that were required for growth on Tf or Lf. Computational modeling of the YfeA homologue, AfeA, revealed conserved residues involved in metal binding.
Organizational Affiliation:
Department of Microbiology, Immunology and Infectious Diseases, Cumming School of Medicine, University of Calgary, Calgary, Canada.