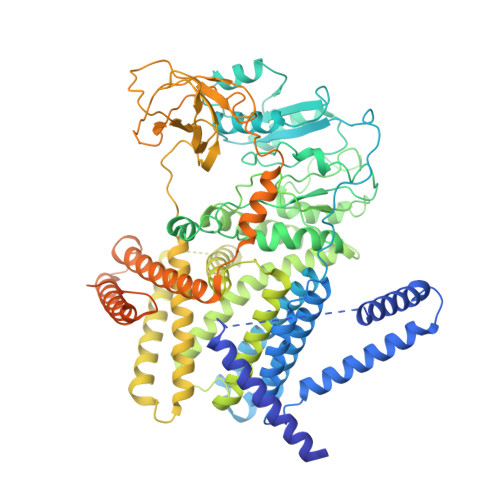
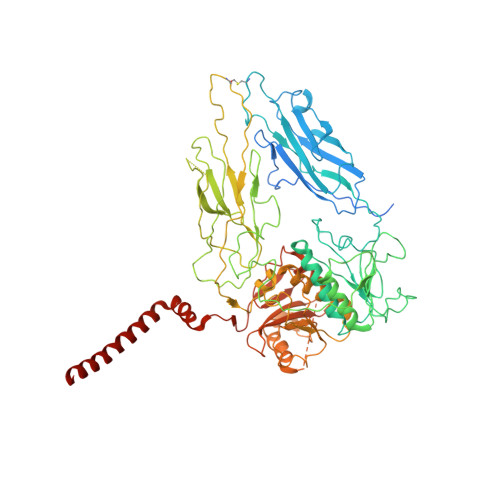
Molecular organization of the E. coli cellulose synthase macrocomplex.
Acheson, J.F., Ho, R., Goularte, N.F., Cegelski, L., Zimmer, J.(2021) Nat Struct Mol Biol 28: 310-318
- PubMed: 33712813
- DOI: https://doi.org/10.1038/s41594-021-00569-7
- Primary Citation of Related Structures:
7L2Z, 7LBY - PubMed Abstract:
Cellulose is frequently found in communities of sessile bacteria called biofilms. Escherichia coli and other enterobacteriaceae modify cellulose with phosphoethanolamine (pEtN) to promote host tissue adhesion. The E. coli pEtN cellulose biosynthesis machinery contains the catalytic BcsA-B complex that synthesizes and secretes cellulose, in addition to five other subunits. The membrane-anchored periplasmic BcsG subunit catalyzes pEtN modification. Here we present the structure of the roughly 1 MDa E. coli Bcs complex, consisting of one BcsA enzyme associated with six copies of BcsB, determined by single-particle cryo-electron microscopy. BcsB homo-oligomerizes primarily through interactions between its carbohydrate-binding domains as well as intermolecular beta-sheet formation. The BcsB hexamer creates a half spiral whose open side accommodates two BcsG subunits, directly adjacent to BcsA's periplasmic channel exit. The cytosolic BcsE and BcsQ subunits associate with BcsA's regulatory PilZ domain. The macrocomplex is a fascinating example of cellulose synthase specification.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia School of Medicine, Charlottesville, VA, USA.