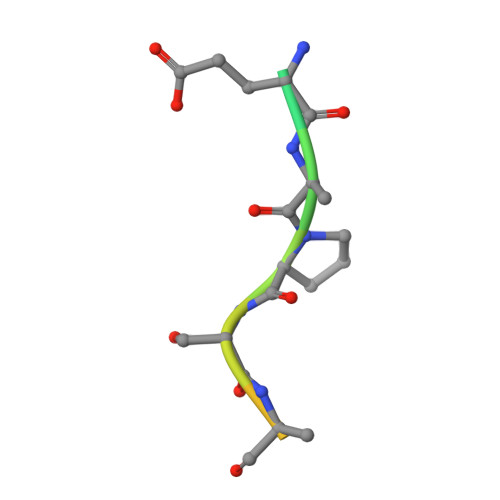
Structural evidence for a proline-specific glycopeptide recognition domain in an O-glycopeptidase.
Noach, I., Boraston, A.B.(2021) Glycobiology 31: 385-390
- PubMed: 33030205
- DOI: https://doi.org/10.1093/glycob/cwaa095
- Primary Citation of Related Structures:
7JTV - PubMed Abstract:
The glycosylation of proteins is typically considered as a stabilizing modification, including resistance to proteolysis. A class of peptidases, referred to as glycopeptidases or O-glycopeptidases, circumvent the protective effect of glycans against proteolysis by accommodating the glycans in their active sites as specific features of substrate recognition. IMPa from Pseudomonas aeruginosa is such an O-glycopeptidase that cleaves the peptide bond immediately preceding a site of O-glycosylation, and through this glycoprotein-degrading function contributes to the host-pathogen interaction. IMPa, however, is a relatively large multidomain protein and how its additional domains may contribute to its function remains unknown. Here, through the determination of a crystal structure of IMPa in complex with an O-glycopeptide, we reveal that the N-terminal domain of IMPa, which is classified in Pfam as IMPa_N_2, is a proline recognition domain that also shows the properties of recognizing an O-linked glycan on the serine/threonine residue following the proline. The proline is bound in the center of a bowl formed by four functionally conserved aromatic amino acid side chains while the glycan wraps around one of the tyrosine residues in the bowl to make classic aromatic ring-carbohydrate CH-π interactions. This structural evidence provides unprecedented insight into how the ancillary domains in glycoprotein-specific peptidases can noncatalytically recognize specific glycosylated motifs that are common in mucin and mucin-like molecules.
Organizational Affiliation:
Biochemistry & Microbiology, University of Victoria, PO Box 3055 STN CSC, Victoria, BC V8W 3P6, Canada.