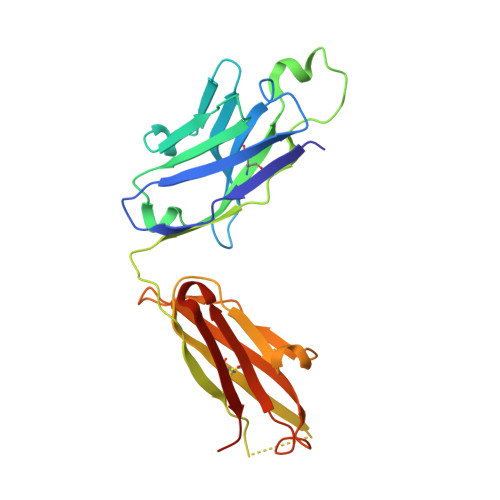
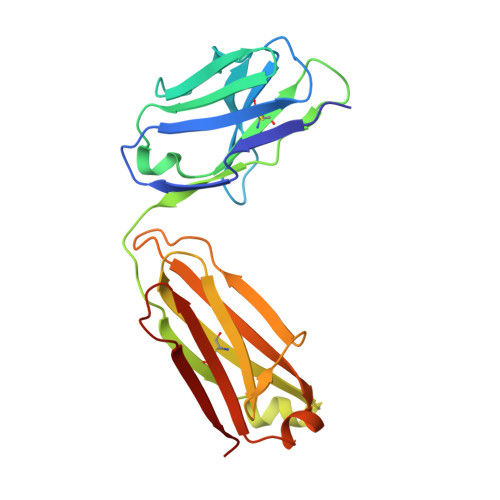
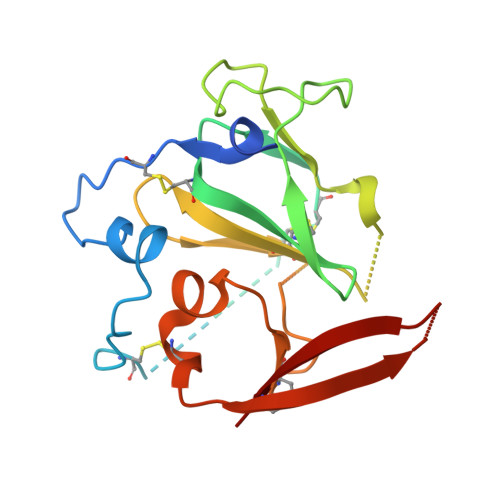
HCV Env immunization elicits a rapid VH1-69-like broadly neutralizing antibody response in rhesus macaques
Tzarum, N., Chen, F., Lin, X., Giang, E., Velazquez-Moctezuma, R., Augestad, E.H., Nagy, K., He, L., Davidson, E., Chavez, D., Prentoe, J., Stanfield, R.L., Lanford, R., Bukh, J., Wilson, I.A., Zhu, J., Law, M.(2021) Immunity