Potent SARS-CoV-2 neutralizing antibodies with protective efficacy against newly emerged mutational variants.
Li, T., Han, X., Gu, C., Guo, H., Zhang, H., Wang, Y., Hu, C., Wang, K., Liu, F., Luo, F., Zhang, Y., Hu, J., Wang, W., Li, S., Hao, Y., Shen, M., Huang, J., Long, Y., Song, S., Wu, R., Mu, S., Chen, Q., Gao, F., Wang, J., Long, S., Li, L., Wu, Y., Gao, Y., Xu, W., Cai, X., Qu, D., Zhang, Z., Zhang, H., Li, N., Gao, Q., Zhang, G., He, C., Wang, W., Ji, X., Tang, N., Yuan, Z., Xie, Y., Yang, H., Zhang, B., Huang, A., Jin, A.(2021) Nat Commun 12: 6304-6304
- PubMed: 34728625
- DOI: https://doi.org/10.1038/s41467-021-26539-7
- Primary Citation of Related Structures:
7E3K, 7E3L - PubMed Abstract:
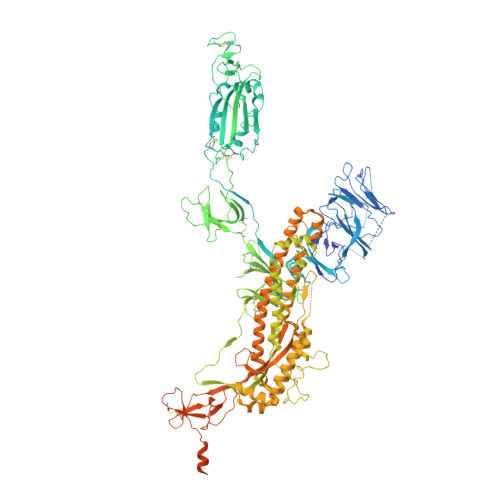
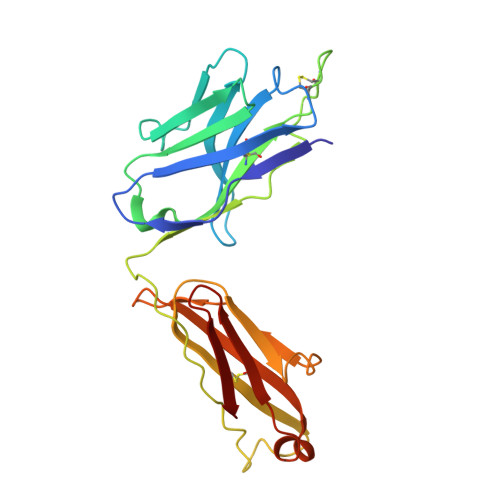
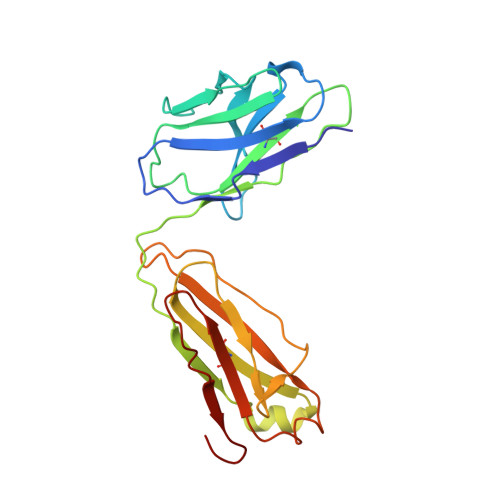
Accumulating mutations in the SARS-CoV-2 Spike (S) protein can increase the possibility of immune escape, challenging the present COVID-19 prophylaxis and clinical interventions. Here, 3 receptor binding domain (RBD) specific monoclonal antibodies (mAbs), 58G6, 510A5 and 13G9, with high neutralizing potency blocking authentic SARS-CoV-2 virus display remarkable efficacy against authentic B.1.351 virus. Surprisingly, structural analysis has revealed that 58G6 and 13G9 both recognize the steric region S 470-495 on the RBD, overlapping the E484K mutation presented in B.1.351. Also, 58G6 directly binds to another region S 450-458 in the RBD. Significantly, 58G6 and 510A5 both demonstrate prophylactic efficacy against authentic SARS-CoV-2 and B.1.351 viruses in the transgenic mice expressing human ACE2 (hACE2), protecting weight loss and reducing virus loads. Together, we have evidenced 2 potent neutralizing Abs with unique mechanism targeting authentic SARS-CoV-2 mutants, which can be promising candidates to fulfill the urgent needs for the prolonged COVID-19 pandemic.
Organizational Affiliation:
Department of Immunology, College of Basic Medicine, Chongqing Medical University, Chongqing, 400010, China.