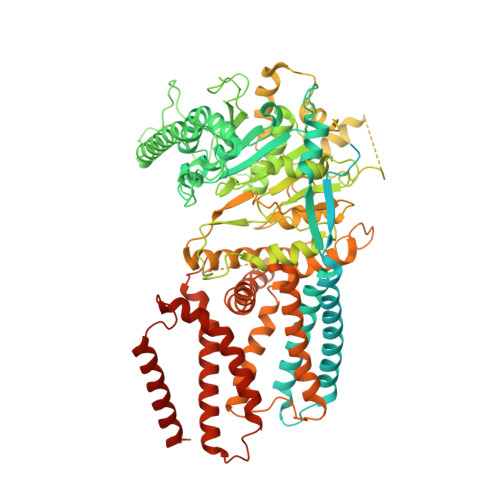
Structural insights into homotrimeric assembly of cellulose synthase CesA7 from Gossypium hirsutum.
Zhang, X., Xue, Y., Guan, Z., Zhou, C., Nie, Y., Men, S., Wang, Q., Shen, C., Zhang, D., Jin, S., Tu, L., Yin, P., Zhang, X.(2021) Plant Biotechnol J 19: 1579-1587
- PubMed: 33638282
- DOI: https://doi.org/10.1111/pbi.13571
- Primary Citation of Related Structures:
7D5K - PubMed Abstract:
Cellulose is one of the most abundant organic polymers in nature. It contains multiple β-1,4-glucan chains synthesized by cellulose synthases (CesAs) on the plasma membrane of higher plants. CesA subunits assemble into a pseudo-sixfold symmetric cellulose synthase complex (CSC), known as a 'rosette complex'. The structure of CesA remains enigmatic. Here, we report the cryo-EM structure of the homotrimeric CesA7 from Gossypium hirsutum at 3.5-angstrom resolution. The GhCesA7 homotrimer shows a C3 symmetrical assembly. Each protomer contains seven transmembrane helices (TMs) which form a channel potentially facilitating the release of newly synthesized glucans. The cytoplasmic glycosyltransferase domain (GT domain) of GhCesA7 protrudes from the membrane, and its catalytic pocket is directed towards the TM pore. The homotrimer GhCesA7 is stabilized by the transmembrane helix 7 (TM7) and the plant-conserved region (PCR) domains. It represents the building block of CSCs and facilitates microfibril formation. This structure provides insight into how eukaryotic cellulose synthase assembles and provides a mechanistic basis for the improvement of cotton fibre quality in the future.
Organizational Affiliation:
National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research, Huazhong Agricultural University, Wuhan, China.