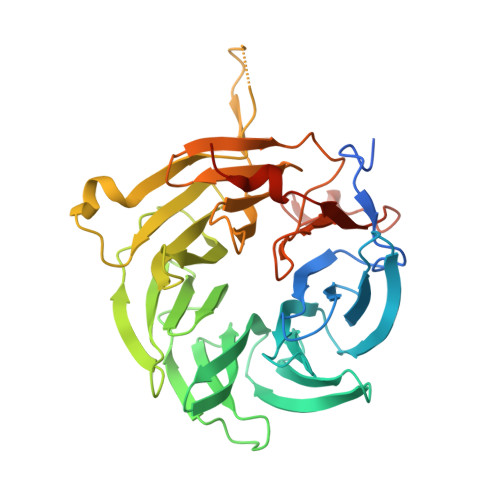
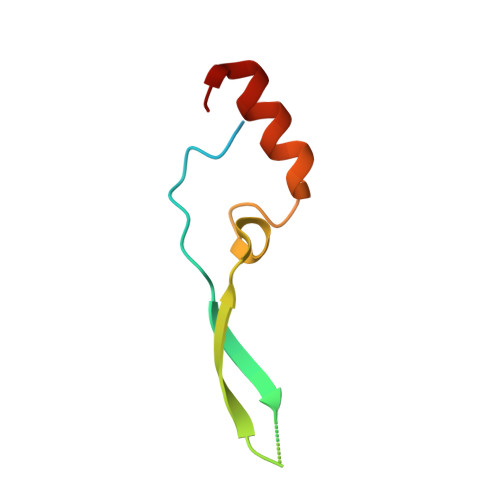
Molecular Mechanism of SARS-CoVs Orf6 Targeting the Rae1-Nup98 Complex to Compete With mRNA Nuclear Export.
Li, T., Wen, Y., Guo, H., Yang, T., Yang, H., Ji, X.(2021) Front Mol Biosci 8: 813248-813248
- PubMed: 35096974
- DOI: https://doi.org/10.3389/fmolb.2021.813248
- Primary Citation of Related Structures:
7VPG, 7VPH - PubMed Abstract:
The accessory protein Orf6 is uniquely expressed in sarbecoviruses including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which is an ongoing pandemic. SARS-CoV-2 Orf6 antagonizes host interferon signaling by inhibition of mRNA nuclear export through its interactions with the ribonucleic acid export 1 (Rae1)-nucleoporin 98 (Nup98) complex. Here, we confirmed the direct tight binding of Orf6 to the Rae1-Nup98 complex, which competitively inhibits RNA binding. We determined the crystal structures of both SARS-CoV-2 and SARS-CoV-1 Orf6 C-termini in complex with the Rae1-Nup98 heterodimer. In each structure, SARS-CoV Orf6 occupies the same potential mRNA-binding groove of the Rae1-Nup98 complex, comparable to the previously reported structures of other viral proteins complexed with Rae1-Nup98, indicating that the Rae1-Nup98 complex is a common target for different viruses to impair the nuclear export pathway. Structural analysis and biochemical studies highlight the critical role of the highly conserved methionine (M58) of SARS-CoVs Orf6. Altogether our data unravel a mechanistic understanding of SARS-CoVs Orf6 targeting the mRNA-binding site of the Rae1-Nup98 complex to compete with the nuclear export of host mRNA, which further emphasizes that Orf6 is a critical virulence factor of SARS-CoVs.
Organizational Affiliation:
The State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Institute of Viruses and Infectious Diseases, Chemistry and Biomedicine Innovation Center (ChemBIC), Institute of Artificial Intelligence Biomedicine, Nanjing University, Nanjing, China.