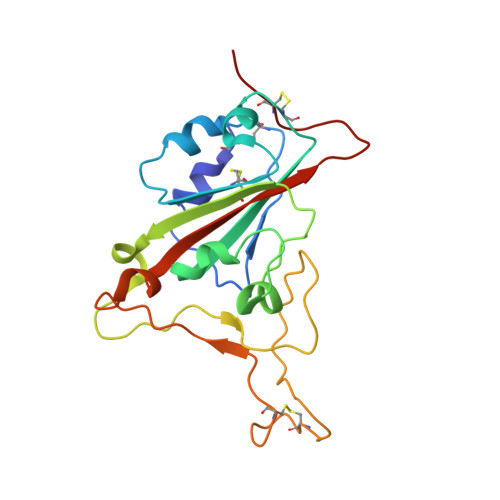
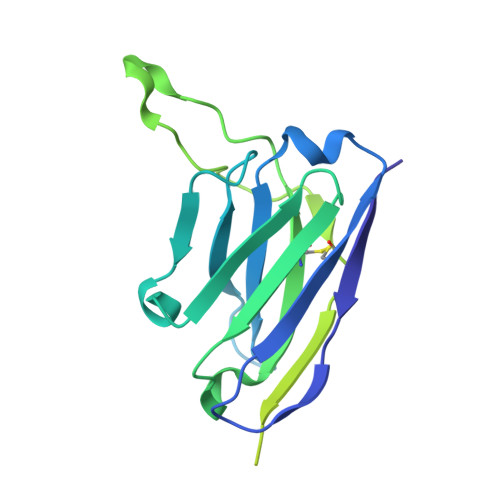
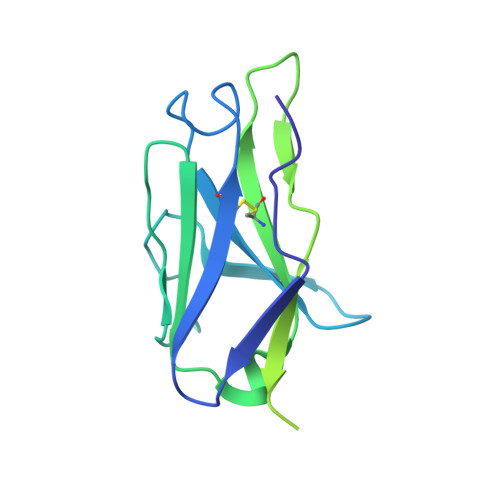
Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Windsor, I.W., Tong, P., Lavidor, O., Moghaddam, A.S., McKay, L.G.A., Gautam, A., Chen, Y., MacDonald, E.A., Yoo, D.K., Griffths, A., Wesemann, D.R., Harrison, S.C.(2022) Sci Immunol 7: eabo3425-eabo3425
- PubMed: 35536154
- DOI: https://doi.org/10.1126/sciimmunol.abo3425
- Primary Citation of Related Structures:
7SWN, 7SWO, 7SWP - PubMed Abstract:
Neutralizing antibodies that recognize the SARS-CoV-2 spike glycoprotein are the principal host defense against viral invasion. Variants of SARS-CoV-2 bear mutations that allow escape from neutralization by many human antibodies, especially those in widely distributed ("public") classes. Identifying antibodies that neutralize these variants of concern and determining their prevalence are important goals for understanding immune protection. To determine the Delta and Omicron BA.1 variant specificity of B cell repertoires established by an initial Wuhan strain infection, we measured neutralization potencies of 73 antibodies from an unbiased survey of the early memory B cell response. Antibodies recognizing each of three previously defined epitopic regions on the spike receptor binding domain (RBD) varied in neutralization potency and variant-escape resistance. The ACE2 binding surface ("RBD-2") harbored the binding sites of neutralizing antibodies with the highest potency but with the greatest sensitivity to viral escape; two other epitopic regions on the RBD ("RBD-1" and "RBD-3") bound antibodies of more modest potency but greater breadth. The structures of several Fab:spike complexes that neutralized all five variants of concern tested, including one Fab each from the RBD-1, -2, and -3 clusters, illustrated the determinants of broad neutralization and showed that B cell repertoires can have specificities that avoid immune escape driven by public antibodies. The structure of the RBD-2 binding, broad neutralizer shows why it retains neutralizing activity for Omicron BA.1, unlike most others in the same public class. Our results correlate with real-world data on vaccine efficacy, which indicate mitigation of disease caused by Omicron BA.1.
Organizational Affiliation:
Boston Children's Hospital, Boston, MA 02115, USA.