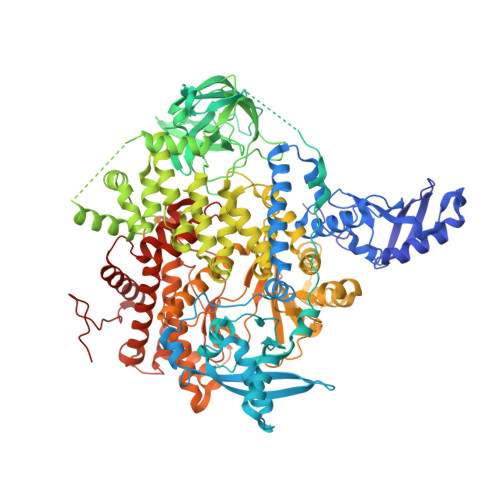
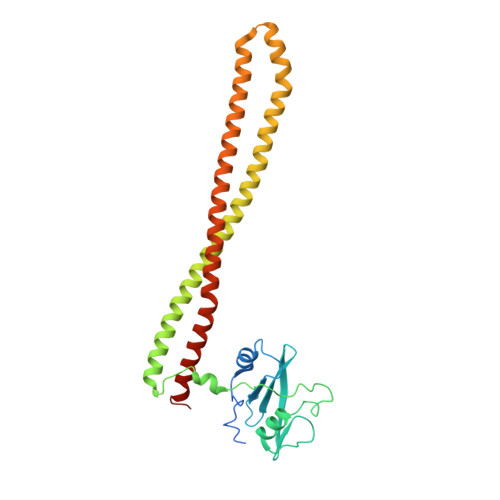
A small-molecule PI3K alpha activator for cardioprotection and neuroregeneration.
Gong, G.Q., Bilanges, B., Allsop, B., Masson, G.R., Roberton, V., Askwith, T., Oxenford, S., Madsen, R.R., Conduit, S.E., Bellini, D., Fitzek, M., Collier, M., Najam, O., He, Z., Wahab, B., McLaughlin, S.H., Chan, A.W.E., Feierberg, I., Madin, A., Morelli, D., Bhamra, A., Vinciauskaite, V., Anderson, K.E., Surinova, S., Pinotsis, N., Lopez-Guadamillas, E., Wilcox, M., Hooper, A., Patel, C., Whitehead, M.A., Bunney, T.D., Stephens, L.R., Hawkins, P.T., Katan, M., Yellon, D.M., Davidson, S.M., Smith, D.M., Phillips, J.B., Angell, R., Williams, R.L., Vanhaesebroeck, B.(2023) Nature 618: 159-168
- PubMed: 37225977
- DOI: https://doi.org/10.1038/s41586-023-05972-2
- Primary Citation of Related Structures:
7PG5, 7PG6, 8BFU, 8OW2 - PubMed Abstract:
Harnessing the potential beneficial effects of kinase signalling through the generation of direct kinase activators remains an underexplored area of drug development 1-5 . This also applies to the PI3K signalling pathway, which has been extensively targeted by inhibitors for conditions with PI3K overactivation, such as cancer and immune dysregulation. Here we report the discovery of UCL-TRO-1938 (referred to as 1938 hereon), a small-molecule activator of the PI3Kα isoform, a crucial effector of growth factor signalling. 1938 allosterically activates PI3Kα through a distinct mechanism by enhancing multiple steps of the PI3Kα catalytic cycle and causes both local and global conformational changes in the PI3Kα structure. This compound is selective for PI3Kα over other PI3K isoforms and multiple protein and lipid kinases. It transiently activates PI3K signalling in all rodent and human cells tested, resulting in cellular responses such as proliferation and neurite outgrowth. In rodent models, acute treatment with 1938 provides cardioprotection from ischaemia-reperfusion injury and, after local administration, enhances nerve regeneration following nerve crush. This study identifies a chemical tool to directly probe the PI3Kα signalling pathway and a new approach to modulate PI3K activity, widening the therapeutic potential of targeting these enzymes through short-term activation for tissue protection and regeneration. Our findings illustrate the potential of activating kinases for therapeutic benefit, a currently largely untapped area of drug development.
Organizational Affiliation:
Cell Signalling, Cancer Institute, University College London, London, UK.