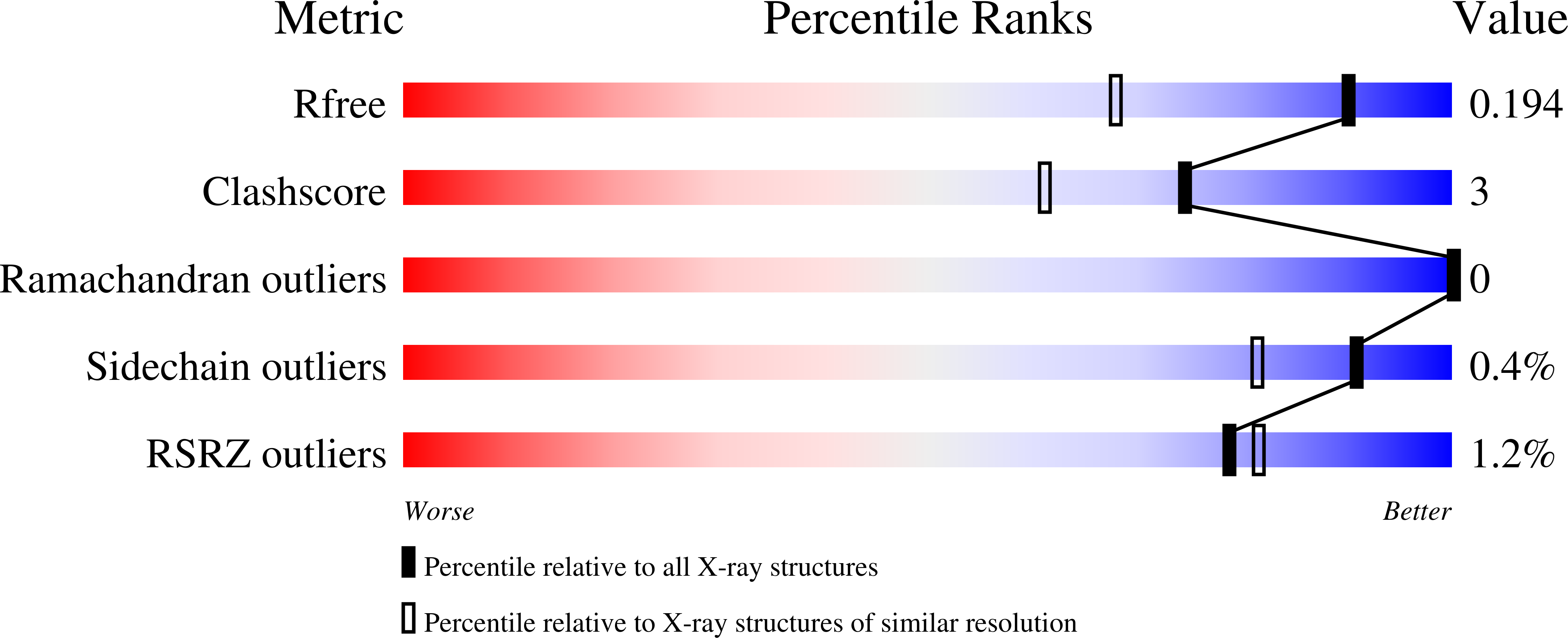
Highlighting the factors governing transglycosylation in the GH5_5 endo-1,4-beta-glucanase RBcel1.
Collet, L., Vander Wauven, C., Oudjama, Y., Galleni, M., Dutoit, R.(2022) Acta Crystallogr D Struct Biol 78: 278-289
- PubMed: 35234142
- DOI: https://doi.org/10.1107/S2059798321013541
- Primary Citation of Related Structures:
7P6G, 7P6H, 7P6I, 7P6J - PubMed Abstract:
Transglycosylating glycoside hydrolases (GHs) offer great potential for the enzymatic synthesis of oligosaccharides. Although knowledge is progressing, there is no unique strategy to improve the transglycosylation yield. Obtaining efficient enzymatic tools for glycan synthesis with GHs remains dependent on an improved understanding of the molecular factors governing the balance between hydrolysis and transglycosylation. This enzymatic and structural study of RBcel1, a transglycosylase from the GH5_5 subfamily isolated from an uncultured bacterium, aims to unravel such factors. The size of the acceptor and donor sugars was found to be critical since transglycosylation is efficient with oligosaccharides at least the size of cellotetraose as the donor and cellotriose as the acceptor. The reaction pH is important in driving the balance between hydrolysis and transglycosylation: hydrolysis is favored at pH values below 8, while transglycosylation becomes the major reaction at basic pH. Solving the structures of two RBcel1 variants, RBcel1_E135Q and RBcel1_Y201F, in complex with ligands has brought to light some of the molecular factors behind transglycosylation. The structure of RBcel1_E135Q in complex with cellotriose allowed a +3 subsite to be defined, in accordance with the requirement for cellotriose as a transglycosylation acceptor. The structure of RBcel1_Y201F has been obtained with several transglycosylation intermediates, providing crystallographic evidence of transglycosylation. The catalytic cleft is filled with (i) donors ranging from cellotriose to cellohexaose in the negative subsites and (ii) cellobiose and cellotriose in the positive subsites. Such a structure is particularly relevant since it is the first structure of a GH5 enzyme in complex with transglycosylation products that has been obtained with neither of the catalytic glutamate residues modified.
Organizational Affiliation:
LABIRIS, 1 Avenue Emile Gryzon, 1070 Brussels, Belgium.