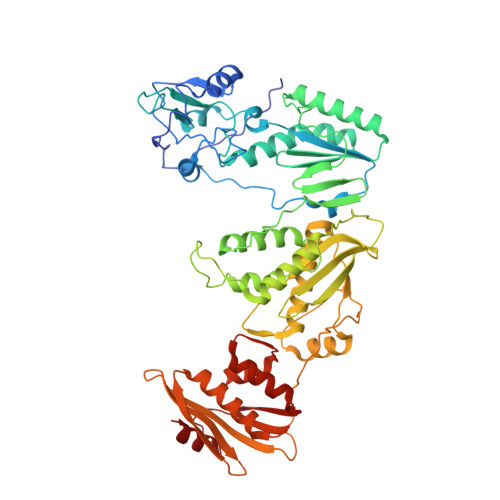
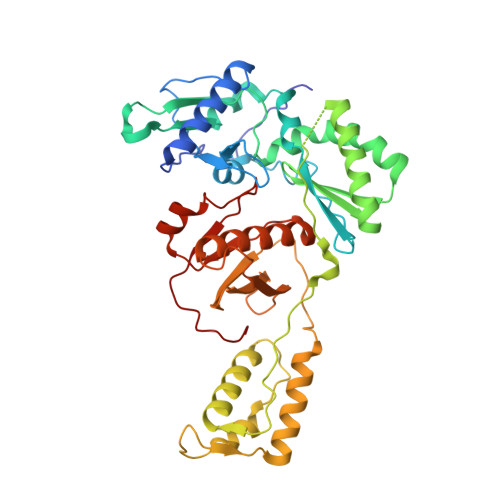
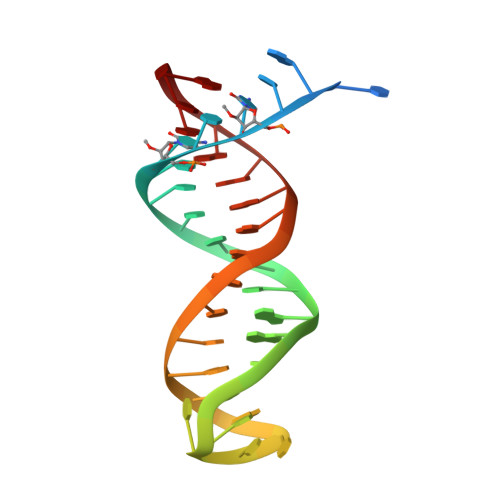
Sliding of HIV-1 reverse transcriptase over DNA creates a transient P pocket - targeting P-pocket by fragment screening.
Singh, A.K., Martinez, S.E., Gu, W., Nguyen, H., Schols, D., Herdewijn, P., De Jonghe, S., Das, K.(2021) Nat Commun 12: 7127-7127
- PubMed: 34880240
- DOI: https://doi.org/10.1038/s41467-021-27409-y
- Primary Citation of Related Structures:
7OXQ, 7OZ2, 7OZ5, 7OZW, 7P15 - PubMed Abstract:
HIV-1 reverse transcriptase (RT) slides over an RNA/DNA or dsDNA substrate while copying the viral RNA to a proviral DNA. We report a crystal structure of RT/dsDNA complex in which RT overstepped the primer 3'-end of a dsDNA substrate and created a transient P-pocket at the priming site. We performed a high-throughput screening of 300 drug-like fragments by X-ray crystallography that identifies two leads that bind the P-pocket, which is composed of structural elements from polymerase active site, primer grip, and template-primer that are resilient to drug-resistance mutations. Analogs of a fragment were synthesized, two of which show noticeable RT inhibition. An engineered RT/DNA aptamer complex could trap the transient P-pocket in solution, and structures of the RT/DNA complex were determined in the presence of an inhibitory fragment. A synthesized analog bound at P-pocket is further analyzed by single-particle cryo-EM. Identification of the P-pocket within HIV RT and the developed structure-based platform provide an opportunity for the design new types of polymerase inhibitors.
Organizational Affiliation:
Department of Microbiology, Immunology and Transplantation, Laboratory of Virology and Chemotherapy, Rega Institute for Medical Research, KU Leuven, Herestraat 49, 3000, Leuven, Belgium.