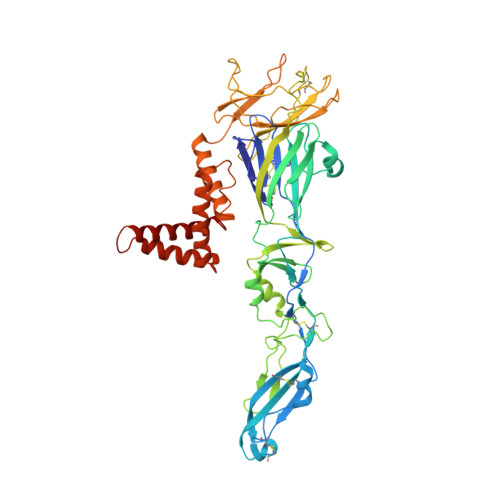
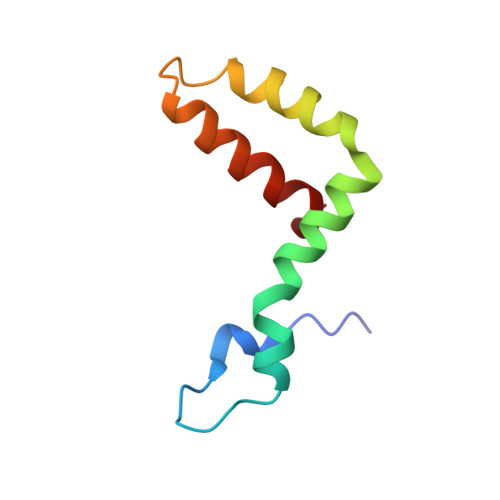
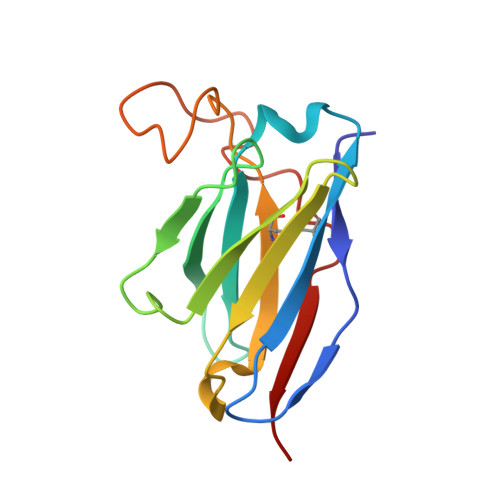
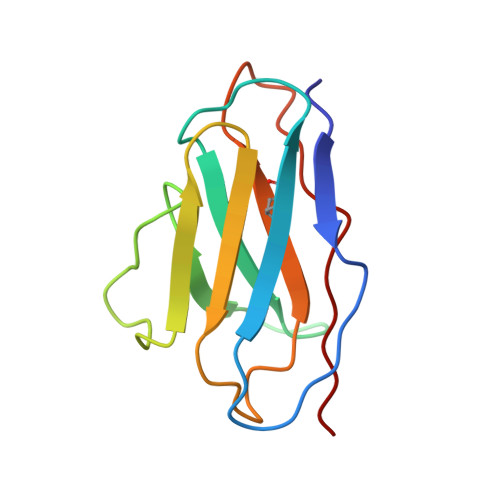
A Human Antibody Neutralizes Different Flaviviruses by Using Different Mechanisms.
Zhang, S., Loy, T., Ng, T.S., Lim, X.N., Chew, S.V., Tan, T.Y., Xu, M., Kostyuchenko, V.A., Tukijan, F., Shi, J., Fink, K., Lok, S.M.(2020) Cell Rep 31: 107584-107584
- PubMed: 32348755
- DOI: https://doi.org/10.1016/j.celrep.2020.107584
- Primary Citation of Related Structures:
7BU8, 7BUA, 7BUB, 7BUD, 7BUE, 7BUF - PubMed Abstract:
Human antibody SIgN-3C neutralizes dengue virus (DENV) and Zika virus (ZIKV) differently. DENV:SIgN-3C Fab and ZIKV:SIgN-3C Fab cryoelectron microscopy (cryo-EM) complex structures show Fabs crosslink E protein dimers at extracellular pH 8.0 condition and also when further incubated at acidic endosomal conditions (pH 8.0-6.5). We observe Fab binding to DENV (pH 8.0-5.0) prevents virus fusion, and the number of bound Fabs increase (from 120 to 180). For ZIKV, although there are already 180 copies of Fab at pH 8.0, virus structural changes at pH 5.0 are not inhibited. The immunoglobulin G (IgG):DENV structure at pH 8.0 shows both Fab arms bind to epitopes around the 2-fold vertex. On ZIKV, an additional Fab around the 5-fold vertex at pH 8.0 suggests one IgG arm would engage with an epitope, although the other may bind to other viruses, causing aggregation. For DENV2 at pH 5.0, a similar scenario would occur, suggesting DENV2:IgG complex would aggregate in the endosome. Hence, a single antibody employs different neutralization mechanisms against different flaviviruses.
Organizational Affiliation:
Program in Emerging Infectious Diseases, Duke-National University of Singapore Medical School, Singapore 169857, Singapore; Centre for BioImaging Sciences, National University of Singapore, Singapore 117557, Singapore.