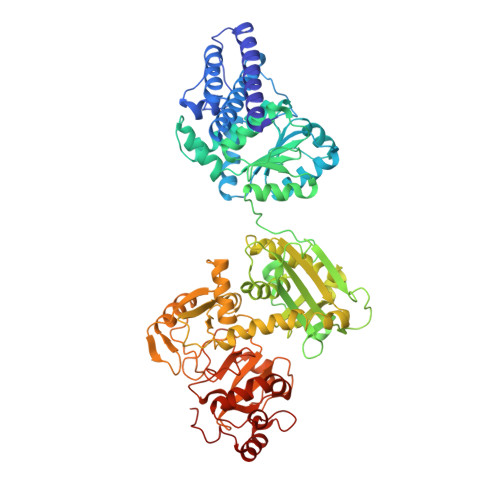
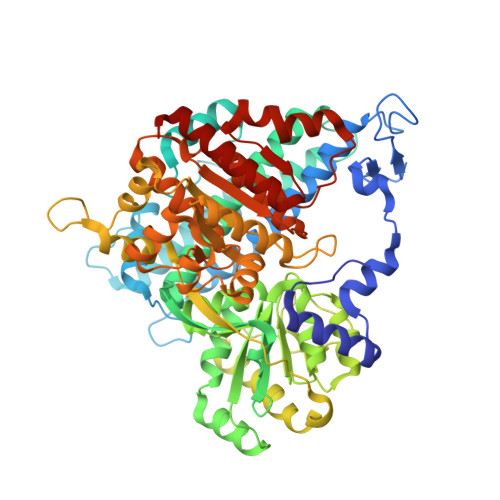
Gas channel rerouting in a primordial enzyme: Structural insights of the carbon-monoxide dehydrogenase/acetyl-CoA synthase complex from the acetogen Clostridium autoethanogenum.
Lemaire, O.N., Wagner, T.(2020) Biochim Biophys Acta Bioenerg 1862: 148330-148330
- PubMed: 33080205
- DOI: https://doi.org/10.1016/j.bbabio.2020.148330
- Primary Citation of Related Structures:
6YTT, 6YU9, 6YUA - PubMed Abstract:
Clostridium autoethanogenum, the bacterial model for biological conversion of waste gases into biofuels, grows under extreme carbon-monoxide (CO) concentrations. The strictly anaerobic bacterium derives its entire cellular energy and carbon from this poisonous gas, therefore requiring efficient molecular machineries for CO-conversion. Here, we structurally and biochemically characterized the key enzyme of the CO-converting metabolism: the CO-dehydrogenase/Acetyl-CoA synthase (CODH/ACS). We obtained crystal structures of natively isolated complexes from fructose-grown and CO-grown C. autoethanogenum cultures. Both contain the same isoforms and if the overall structure adopts the classic α 2 β 2 architecture, comparable to the model enzyme from Moorella thermoacetica, the ACS binds a different position on the CODH core. The structural characterization of a proteolyzed complex and the conservation of the binding interface in close homologs rejected the possibility of a crystallization artefact. Therefore, the internal CO-channeling system, critical to transfer CO generated at the C-cluster to the ACS active site, drastically differs in the complex from C. autoethanogenum. The 1.9-Å structure of the CODH alone provides an accurate picture of the new CO-routes, leading to the ACS core and reaching the surface. Increased gas accessibility would allow the simultaneous CO-oxidation and acetyl-CoA production. Biochemical experiments showed higher flexibility of the ACS subunit from C. autoethanogenum compared to M. thermoacetica, albeit monitoring similar CO-oxidation and formation rates. These results show a reshuffling of internal CO-tunnels during evolution of these Firmicutes, putatively leading to a bidirectional complex that ensure a high flux of CO-conversion toward energy conservation, acting as the main cellular powerplant.
Organizational Affiliation:
Max Planck Institute for Marine Microbiology, Celsiusstraße 1, 28359 Bremen, Germany.