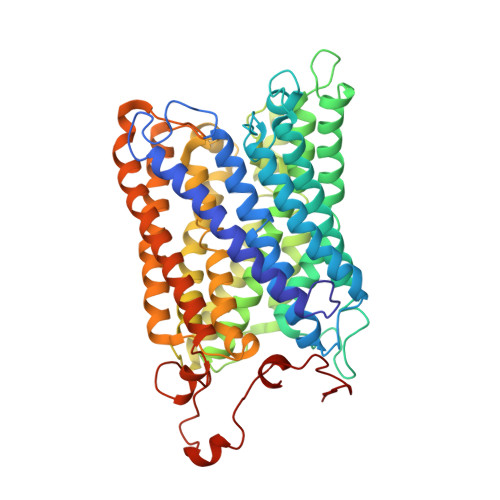
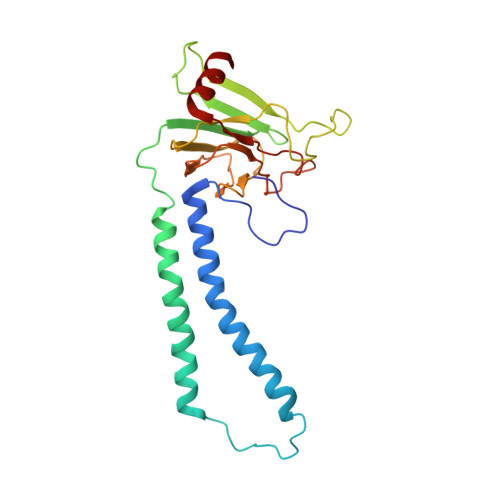
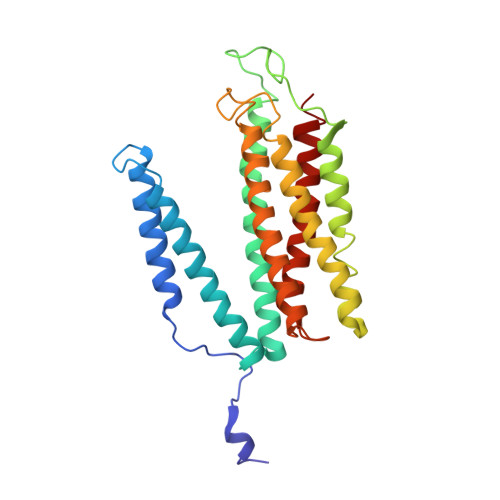
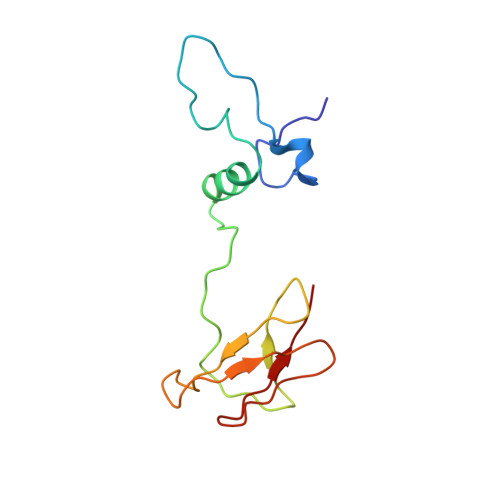
Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Berndtsson, J., Aufschnaiter, A., Rathore, S., Marin-Buera, L., Dawitz, H., Diessl, J., Kohler, V., Barrientos, A., Buttner, S., Fontanesi, F., Ott, M.(2020) EMBO Rep 21: e51015-e51015
- PubMed: 33016568
- DOI: https://doi.org/10.15252/embr.202051015
- Primary Citation of Related Structures:
6YMX, 6YMY - PubMed Abstract:
Respiratory chains are crucial for cellular energy conversion and consist of multi-subunit complexes that can assemble into supercomplexes. These structures have been intensively characterized in various organisms, but their physiological roles remain unclear. Here, we elucidate their function by leveraging a high-resolution structural model of yeast respiratory supercomplexes that allowed us to inhibit supercomplex formation by mutation of key residues in the interaction interface. Analyses of a mutant defective in supercomplex formation, which still contains fully functional individual complexes, show that the lack of supercomplex assembly delays the diffusion of cytochrome c between the separated complexes, thus reducing electron transfer efficiency. Consequently, competitive cellular fitness is severely reduced in the absence of supercomplex formation and can be restored by overexpression of cytochrome c. In sum, our results establish how respiratory supercomplexes increase the efficiency of cellular energy conversion, thereby providing an evolutionary advantage for aerobic organisms.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Stockholm University, Stockholm, Sweden.