Elucidating the 3D Structure of a Surface Membrane Antigen from Trypanosoma cruzi as a Serodiagnostic Biomarker of Chagas Disease.
Di Pisa, F., De Benedetti, S., Fassi, E.M.A., Bombaci, M., Grifantini, R., Musico, A., Frigerio, R., Pontillo, A., Rigo, C., Abelli, S., Grande, R., Zanchetta, N., Mileto, D., Mancon, A., Rizzo, A., Gori, A., Cretich, M., Colombo, G., Bolognesi, M., Gourlay, L.J.(2022) Vaccines (Basel) 10
- PubMed: 35062732
- DOI: https://doi.org/10.3390/vaccines10010071
- Primary Citation of Related Structures:
6Y0D - PubMed Abstract:
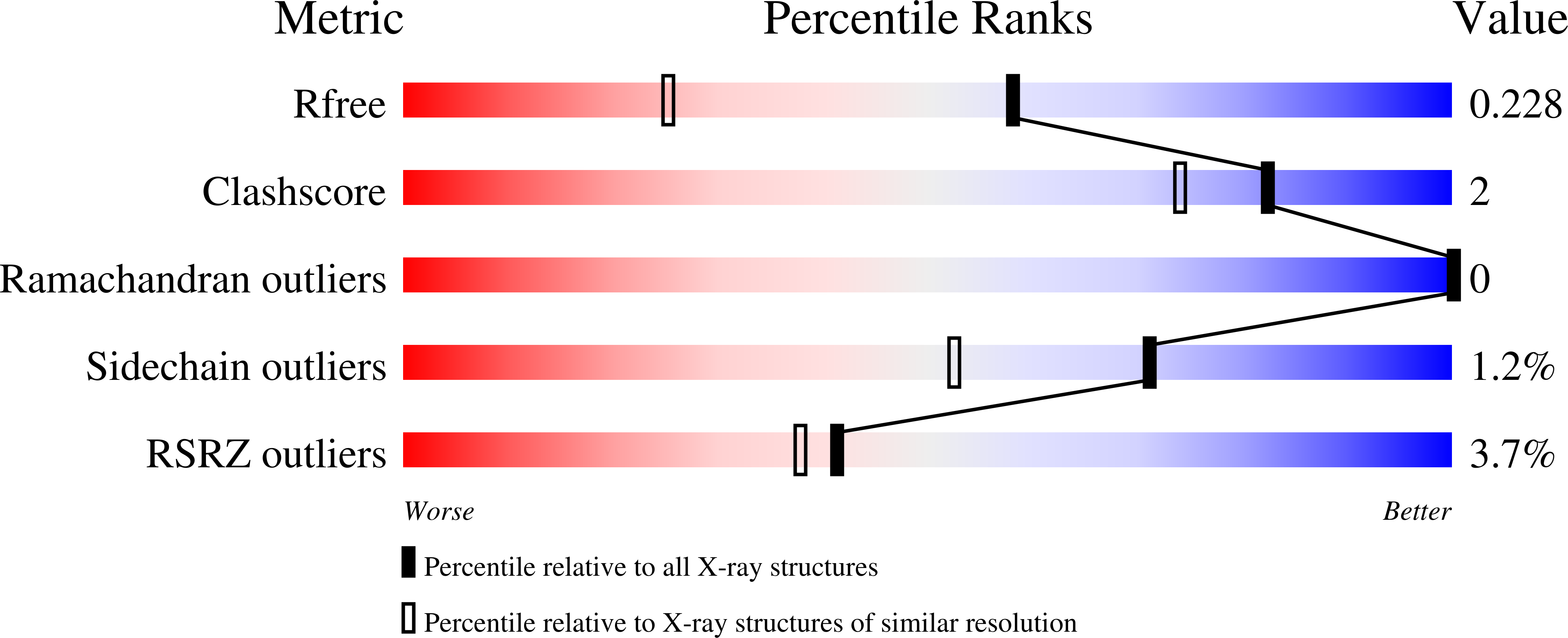
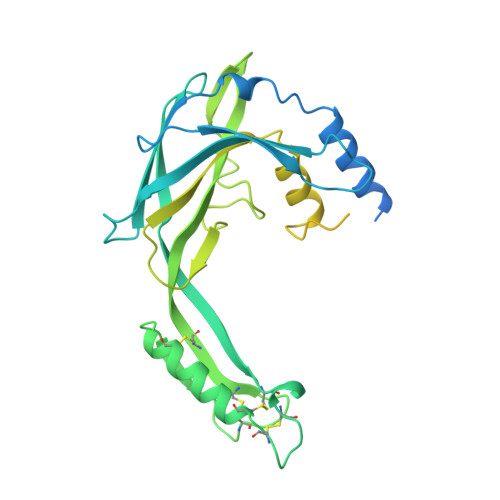
Chagas disease (CD) is a vector-borne parasitosis, caused by the protozoan parasite Trypanosoma cruzi , that affects millions of people worldwide. Although endemic in South America, CD is emerging throughout the world due to climate change and increased immigratory flux of infected people to non-endemic regions. Containing of the diffusion of CD is challenged by the asymptomatic nature of the disease in early infection stages and by the lack of a rapid and effective diagnostic test. With the aim of designing new serodiagnostic molecules to be implemented in a microarray-based diagnostic set-up for early screening of CD, herein, we report the recombinant production of the extracellular domain of a surface membrane antigen from T. cruzi ( Tc SMP) and confirm its ability to detect plasma antibodies from infected patients. Moreover, we describe its high-resolution (1.62 Å) crystal structure, to which in silico epitope predictions were applied in order to locate the most immunoreactive regions of Tc SMP in order to guide the design of epitopes that may be used as an alternative to the full-length antigen for CD diagnosis. Two putative, linear epitopes, belonging to the same immunogenic region, were synthesized as free peptides, and their immunological properties were tested in vitro. Although both peptides were shown to adopt a structural conformation that allowed their recognition by polyclonal antibodies raised against the recombinant protein, they were not serodiagnostic for T. cruzi infections. Nevertheless, they represent good starting points for further iterative structure-based (re)design cycles.
Organizational Affiliation:
Department of Biosciences, Università degli Studi di Milano, Via Celoria 26, 20133 Milano, Italy.