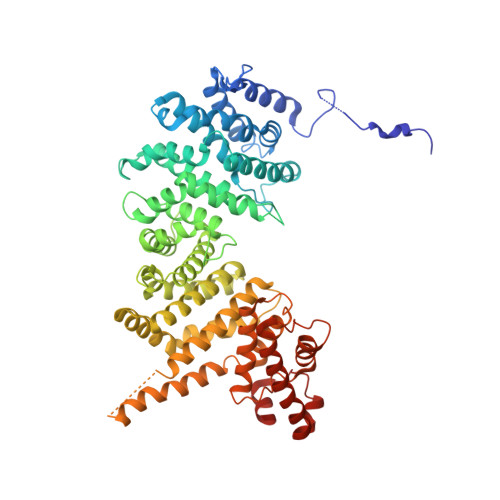
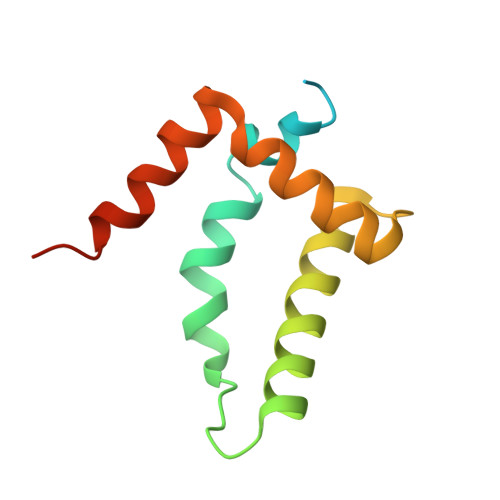
Crystal structure and interactions of the Tof1-Csm3 (Timeless-Tipin) fork protection complex.
Grabarczyk, D.B.(2020) Nucleic Acids Res 48: 6996-7004
- PubMed: 32469068
- DOI: https://doi.org/10.1093/nar/gkaa456
- Primary Citation of Related Structures:
6XWX - PubMed Abstract:
The Tof1-Csm3 fork protection complex has a central role in the replisome-it promotes the progression of DNA replication forks and protects them when they stall, while also enabling cohesion establishment and checkpoint responses. Here, I present the crystal structure of the Tof1-Csm3 complex from Chaetomium thermophilum at 3.1 Å resolution. The structure reveals that both proteins together form an extended alpha helical repeat structure, which suggests a mechanical or scaffolding role for the complex. Expanding on this idea, I characterize a DNA interacting region and a cancer-associated Mrc1 binding site. This study provides the molecular basis for understanding the functions of the Tof1-Csm3 complex, its human orthologue the Timeless-Tipin complex and additionally the Drosophila circadian rhythm protein Timeless.
Organizational Affiliation:
Rudolf Virchow Center for Experimental Biomedicine, Institute for Structural Biology, University of Würzburg, Josef-Schneider-Str. 2, Würzburg, Germany.