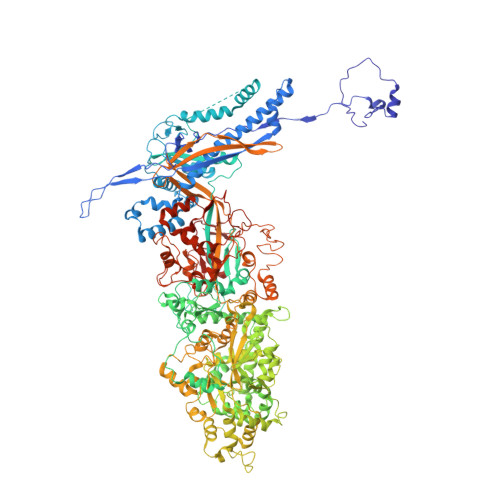
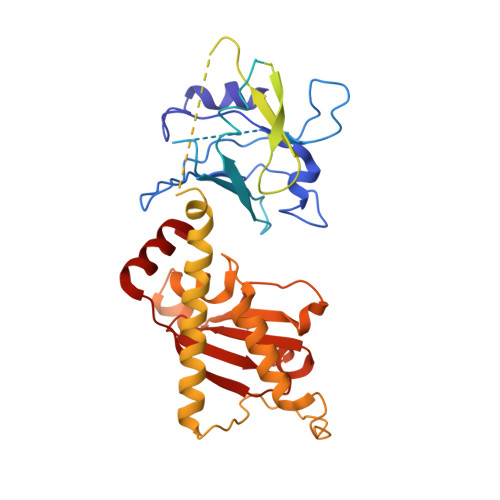
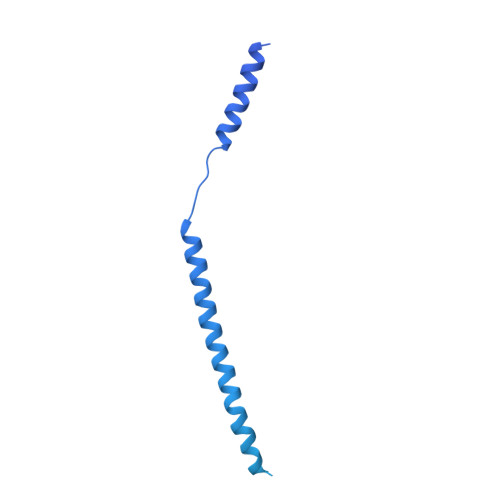
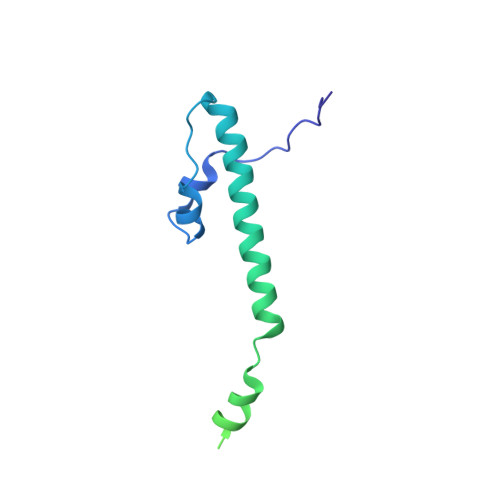
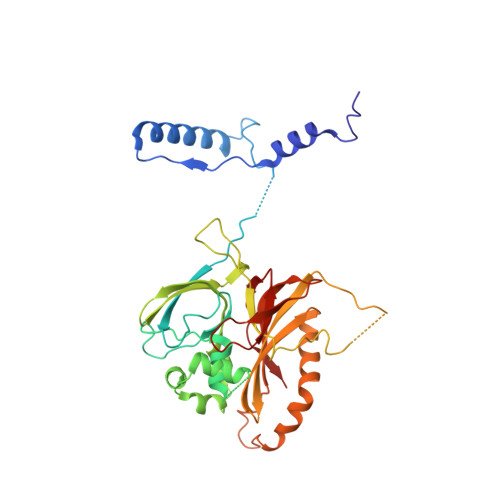
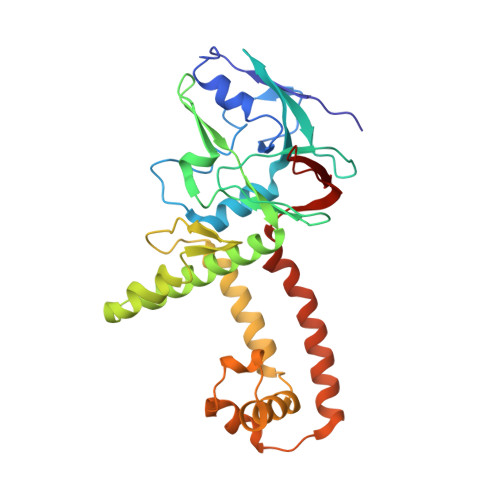
Structures of capsid and capsid-associated tegument complex inside the Epstein-Barr virus.
Liu, W., Cui, Y., Wang, C., Li, Z., Gong, D., Dai, X., Bi, G.Q., Sun, R., Zhou, Z.H.(2020) Nat Microbiol 5: 1285-1298
- PubMed: 32719506
- DOI: https://doi.org/10.1038/s41564-020-0758-1
- Primary Citation of Related Structures:
6W19, 6W2D, 6W2E - PubMed Abstract:
As the first discovered human cancer virus, Epstein-Barr virus (EBV) causes Burkitt's lymphoma and nasopharyngeal carcinoma. Isolating virions for determining high-resolution structures has been hindered by latency-a hallmark of EBV infection-and atomic structures are thus available only for recombinantly expressed EBV proteins. In the present study, by symmetry relaxation and subparticle reconstruction, we have determined near-atomic-resolution structures of the EBV capsid with an asymmetrically attached DNA-translocating portal and capsid-associated tegument complexes from cryogenic electron microscopy images of just 2,048 EBV virions obtained by chemical induction. The resulting atomic models reveal structural plasticity among the 20 conformers of the major capsid protein, 2 conformers of the small capsid protein (SCP), 4 conformers of the triplex monomer proteins and 2 conformers of the triplex dimer proteins. Plasticity reaches the greatest level at the capsid-tegument interfaces involving SCP and capsid-associated tegument complexes (CATC): SCPs crown pentons/hexons and mediate tegument protein binding, and CATCs bind and rotate all five periportal triplexes, but notably only about one peri-penton triplex. These results offer insights into the EBV capsid assembly and a mechanism for recruiting cell-regulating factors into the tegument compartment as 'cargoes', and should inform future anti-EBV strategies.
Organizational Affiliation:
California NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA, USA.