The essential function of ISCU2 and its conserved N-terminus in Fe/S cluster biogenesis
Freibert, S.A., Boniecki, M.T., Shulz, V., Wilbrecht, C., Krapoth, N., Muhlenhoff, U., Stehling, O., Cygler, M., Lill, R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Currently 6UXE does not have a validation slider image.
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cysteine desulfurase, mitochondrial | 406 | Homo sapiens | Mutation(s): 0 Gene Names: NFS1, NIFS, HUSSY-08 EC: 2.8.1.7 | 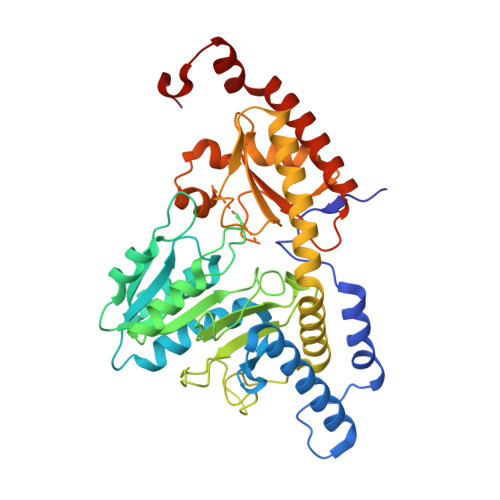 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9Y697 (Homo sapiens) Explore Q9Y697 Go to UniProtKB: Q9Y697 | |||||
PHAROS: Q9Y697 GTEx: ENSG00000244005 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9Y697 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| LYR motif-containing protein 4 | 91 | Homo sapiens | Mutation(s): 1 Gene Names: LYRM4, C6orf149, ISD11, CGI-203 | 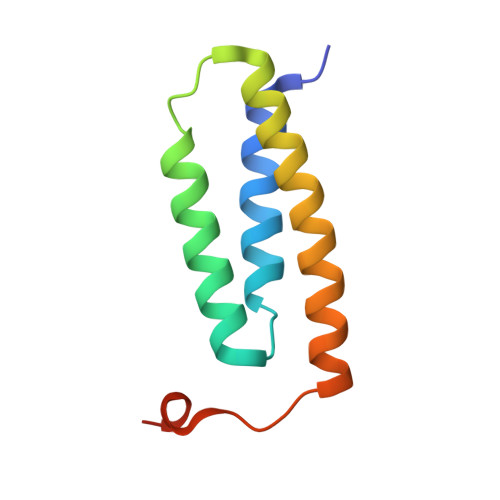 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9HD34 (Homo sapiens) Explore Q9HD34 Go to UniProtKB: Q9HD34 | |||||
PHAROS: Q9HD34 GTEx: ENSG00000214113 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9HD34 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Acyl carrier protein | 77 | Escherichia coli | Mutation(s): 0 Gene Names: acpP, ECS88_1108 | 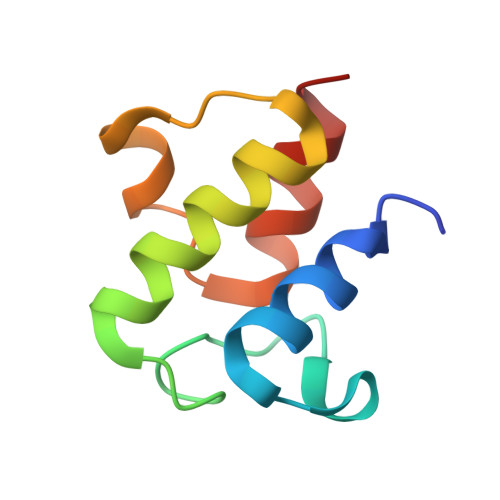 | |
UniProt | |||||
Find proteins for P0A6A8 (Escherichia coli (strain K12)) Explore P0A6A8 Go to UniProtKB: P0A6A8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A6A8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Iron-sulfur cluster assembly enzyme ISCU, mitochondrial | 143 | Homo sapiens | Mutation(s): 1 Gene Names: ISCU, NIFUN | 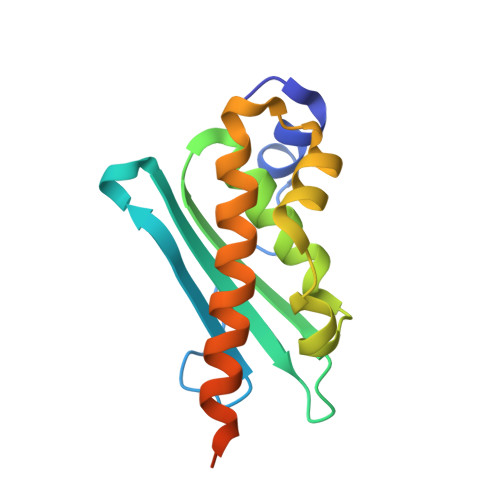 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q9H1K1 (Homo sapiens) Explore Q9H1K1 Go to UniProtKB: Q9H1K1 | |||||
PHAROS: Q9H1K1 GTEx: ENSG00000136003 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9H1K1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 12 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 8Q1 (Subject of Investigation/LOI) Query on 8Q1 | SB [auth C] | S-[2-({N-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] dodecanethioate C23 H45 N2 O8 P S MVHUOSAYFQKAMT-NRFANRHFSA-N |  | ||
| P15 Query on P15 | DA [auth A] | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL C13 H28 O7 FHHGCKHKTAJLOM-UHFFFAOYSA-N |  | ||
| EDT Query on EDT | PB [auth B] | {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID C10 H16 N2 O8 KCXVZYZYPLLWCC-UHFFFAOYSA-N |  | ||
| PLP (Subject of Investigation/LOI) Query on PLP | E [auth A] | PYRIDOXAL-5'-PHOSPHATE C8 H10 N O6 P NGVDGCNFYWLIFO-UHFFFAOYSA-N |  | ||
| 1PE Query on 1PE | AC [auth D] | PENTAETHYLENE GLYCOL C10 H22 O6 JLFNLZLINWHATN-UHFFFAOYSA-N |  | ||
| MES Query on MES | TB [auth C] | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID C6 H13 N O4 S SXGZJKUKBWWHRA-UHFFFAOYSA-N |  | ||
| PG4 Query on PG4 | W [auth A], X [auth A] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| DTT Query on DTT | KA [auth A] | 2,3-DIHYDROXY-1,4-DITHIOBUTANE C4 H10 O2 S2 VHJLVAABSRFDPM-IMJSIDKUSA-N |  | ||
| PGE Query on PGE | LA [auth A], UA [auth A], VA [auth A] | TRIETHYLENE GLYCOL C6 H14 O4 ZIBGPFATKBEMQZ-UHFFFAOYSA-N |  | ||
| PEG Query on PEG | AB [auth A] BB [auth A] EA [auth A] IA [auth A] JA [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| GOL Query on GOL | CB [auth A] CC [auth D] F [auth A] G [auth A] H [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | AA [auth A] BA [auth A] BC [auth D] CA [auth A] DB [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 86.41 | α = 90 |
| b = 86.41 | β = 90 |
| c = 245.79 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
Currently 6UXE does not have a validation slider image.