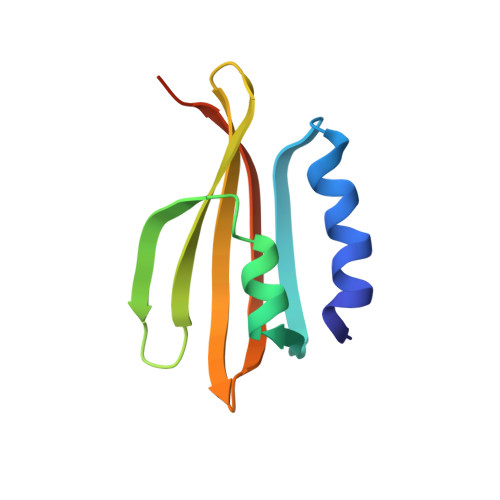
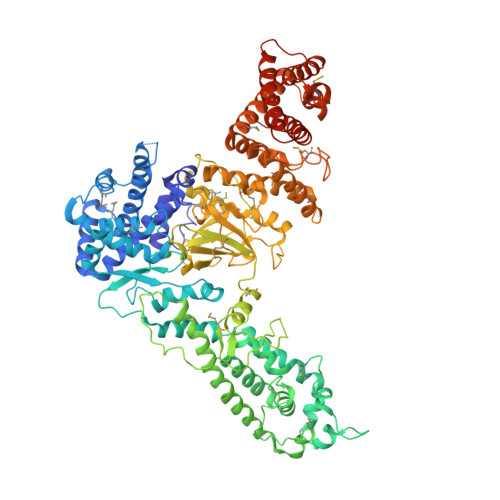
Structural basis for inhibition of an archaeal CRISPR-Cas type I-D large subunit by an anti-CRISPR protein.
Manav, M.C., Van, L.B., Lin, J., Fuglsang, A., Peng, X., Brodersen, D.E.(2020) Nat Commun 11: 5993-5993
- PubMed: 33239638
- DOI: https://doi.org/10.1038/s41467-020-19847-x
- Primary Citation of Related Structures:
6THH, 6YES - PubMed Abstract:
A hallmark of type I CRISPR-Cas systems is the presence of Cas3, which contains both the nuclease and helicase activities required for DNA cleavage during interference. In subtype I-D systems, however, the histidine-aspartate (HD) nuclease domain is encoded as part of a Cas10-like large effector complex subunit and the helicase activity in a separate Cas3' subunit, but the functional and mechanistic consequences of this organisation are not currently understood. Here we show that the Sulfolobus islandicus type I-D Cas10d large subunit exhibits an unusual domain architecture consisting of a Cas3-like HD nuclease domain fused to a degenerate polymerase fold and a C-terminal domain structurally similar to Cas11. Crystal structures of Cas10d both in isolation and bound to S. islandicus rod-shaped virus 3 AcrID1 reveal that the anti-CRISPR protein sequesters the large subunit in a non-functional state unable to form a cleavage-competent effector complex. The architecture of Cas10d suggests that the type I-D effector complex is similar to those found in type III CRISPR-Cas systems and that this feature is specifically exploited by phages for anti-CRISPR defence.
Organizational Affiliation:
Department of Molecular Biology and Genetics, Aarhus University, Gustav Wieds Vej 10c, DK-8000, Aarhus C, Denmark.