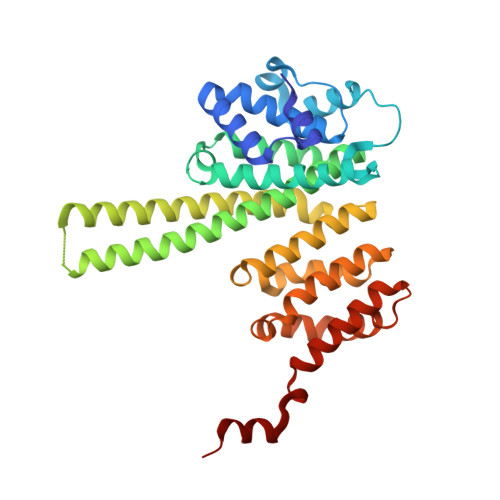
STAG1 vulnerabilities for exploiting cohesin synthetic lethality in STAG2-deficient cancers.
van der Lelij, P., Newman, J.A., Lieb, S., Jude, J., Katis, V., Hoffmann, T., Hinterndorfer, M., Bader, G., Kraut, N., Pearson, M.A., Peters, J.M., Zuber, J., Gileadi, O., Petronczki, M.(2020) Life Sci Alliance 3
- PubMed: 32467316
- DOI: https://doi.org/10.26508/lsa.202000725
- Primary Citation of Related Structures:
6QB5, 6R7O, 6RRC, 6RRK - PubMed Abstract:
The cohesin subunit STAG2 has emerged as a recurrently inactivated tumor suppressor in human cancers. Using candidate approaches, recent studies have revealed a synthetic lethal interaction between STAG2 and its paralog STAG1 To systematically probe genetic vulnerabilities in the absence of STAG2, we have performed genome-wide CRISPR screens in isogenic cell lines and identified STAG1 as the most prominent and selective dependency of STAG2-deficient cells. Using an inducible degron system, we show that chemical genetic degradation of STAG1 protein results in the loss of sister chromatid cohesion and rapid cell death in STAG2-deficient cells, while sparing STAG2 -wild-type cells. Biochemical assays and X-ray crystallography identify STAG1 regions that interact with the RAD21 subunit of the cohesin complex. STAG1 mutations that abrogate this interaction selectively compromise the viability of STAG2 -deficient cells. Our work highlights the degradation of STAG1 and inhibition of its interaction with RAD21 as promising therapeutic strategies. These findings lay the groundwork for the development of STAG1-directed small molecules to exploit synthetic lethality in STAG2 -mutated tumors.
Organizational Affiliation:
Research Institute of Molecular Pathology (IMP), Vienna BioCenter (VBC), Vienna, Austria.