Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Singh, K., Graf, B., Linden, A., Sautner, V., Urlaub, H., Tittmann, K., Stark, H., Chari, A.(2020) Cell 180: 1130-1143.e20
- PubMed: 32160528
- DOI: https://doi.org/10.1016/j.cell.2020.02.034
- Primary Citation of Related Structures:
6QL5, 6QL6, 6QL7, 6QL9 - PubMed Abstract:
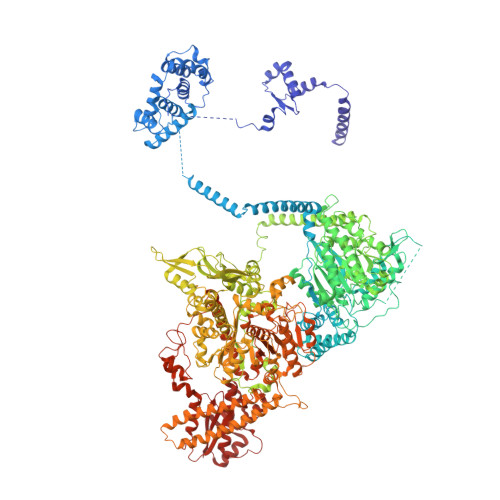
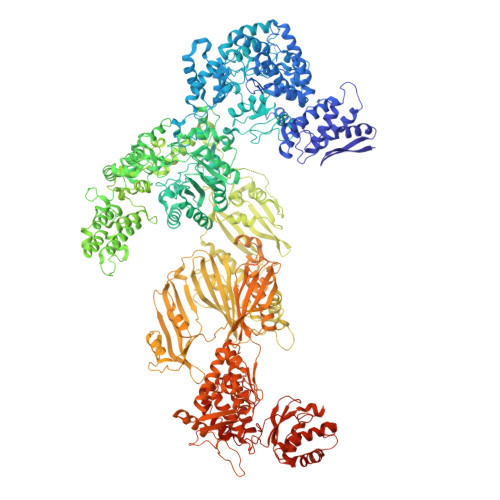
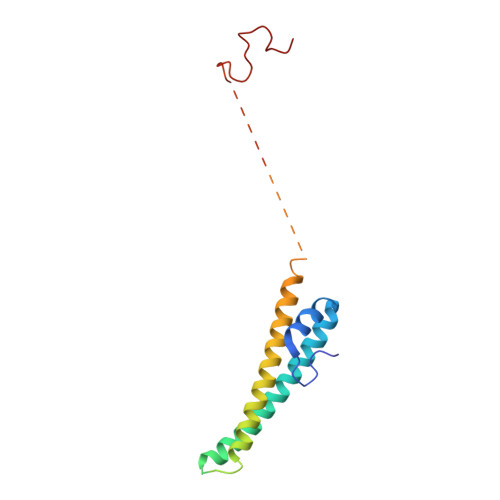
Fatty acid synthases (FASs) are central to metabolism but are also of biotechnological interest for the production of fine chemicals and biofuels from renewable resources. During fatty acid synthesis, the growing fatty acid chain is thought to be shuttled by the dynamic acyl carrier protein domain to several enzyme active sites. Here, we report the discovery of a γ subunit of the 2.6 megadalton α 6 -β 6 S. cerevisiae FAS, which is shown by high-resolution structures to stabilize a rotated FAS conformation and rearrange ACP domains from equatorial to axial positions. The γ subunit spans the length of the FAS inner cavity, impeding reductase activities of FAS, regulating NADPH turnover by kinetic hysteresis at the ketoreductase, and suppressing off-pathway reactions at the enoylreductase. The γ subunit delineates the functional compartment within FAS. As a scaffold, it may be exploited to incorporate natural and designed enzymatic activities that are not present in natural FAS.
Organizational Affiliation:
Department of Structural Dynamics, Max Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany.