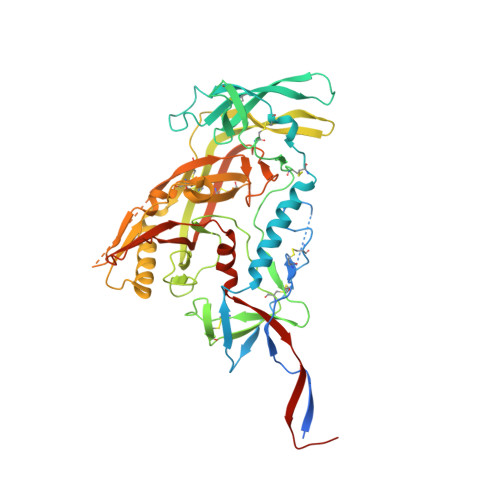
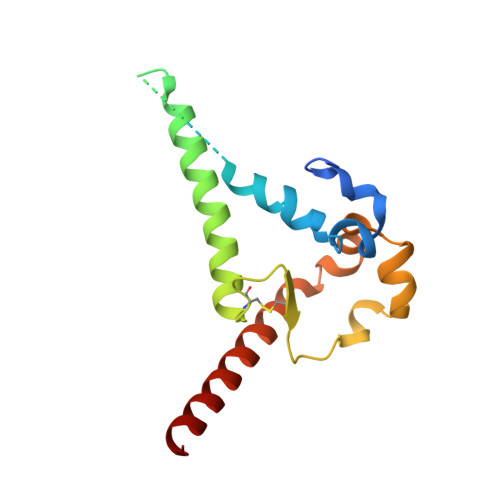
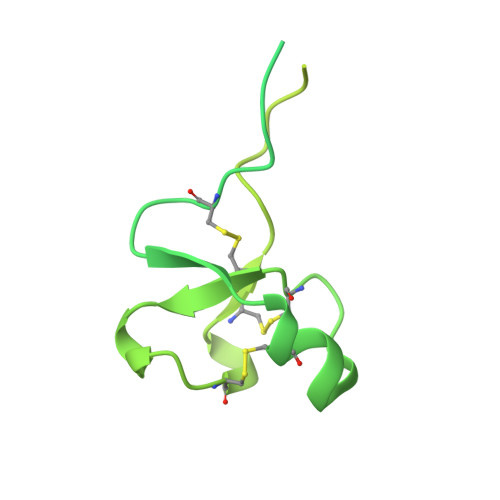
Structural basis of broad HIV neutralization by a vaccine-induced cow antibody.
Stanfield, R.L., Berndsen, Z.T., Huang, R., Sok, D., Warner, G., Torres, J.L., Burton, D.R., Ward, A.B., Wilson, I.A., Smider, V.V.(2020) Sci Adv 6: eaba0468-eaba0468
- PubMed: 32518821
- DOI: https://doi.org/10.1126/sciadv.aba0468
- Primary Citation of Related Structures:
6OO0, 6OPA, 6PW6 - PubMed Abstract:
Potent broadly neutralizing antibodies (bnAbs) to HIV have been very challenging to elicit by vaccination in wild-type animals. Here, by x-ray crystallography, cryo-electron microscopy, and site-directed mutagenesis, we structurally and functionally elucidate the mode of binding of a potent bnAb (NC-Cow1) elicited in cows by immunization with the HIV envelope (Env) trimer BG505 SOSIP.664. The exceptionally long (60 residues) third complementarity-determining region of the heavy chain (CDR H3) of NC-Cow1 forms a mini domain (knob) on an extended stalk that navigates through the dense glycan shield on Env to target a small footprint on the gp120 CD4 receptor binding site with no contact of the other CDRs to the rest of the Env trimer. These findings illustrate, in molecular detail, how an unusual vaccine-induced cow bnAb to HIV Env can neutralize with high potency and breadth.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA 92037, USA.