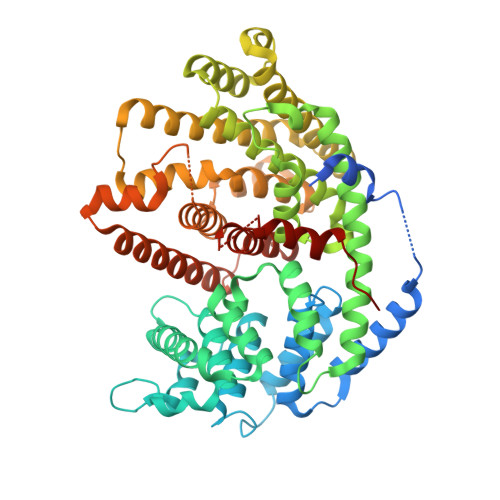
Structure of Sesquisabinene Synthase 1, a Terpenoid Cyclase That Generates a Strained [3.1.0] Bridged-Bicyclic Product.
Blank, P.N., Shinsky, S.A., Christianson, D.W.(2019) ACS Chem Biol 14: 1011-1019
- PubMed: 30977996
- DOI: https://doi.org/10.1021/acschembio.9b00218
- Primary Citation of Related Structures:
6O9P, 6O9Q - PubMed Abstract:
The natural product sesquisabinene is a key component of the fragrant essential oil of the sandalwood tree, currently valued at $5,000/L. Sesquisabinene contains a highly strained [3.1.0] bicyclic ring system and is generated from farnesyl diphosphate in a reaction catalyzed by a class I terpenoid cyclase. To understand how the enzyme directs the formation of a strained hydrocarbon ring system, we now report the X-ray crystal structure of sesquisabinene synthase 1 (SQS1) from the Indian sandalwood tree ( Santalum album). Specifically, we report the structure of unliganded SQS1 at 1.90 Å resolution and the structure of its complex with three Mg 2+ ions and the inhibitor ibandronate at 2.10 Å resolution. The bisphosphonate group of ibandronate coordinates to all three metal ions and makes hydrogen bond interactions with basic residues at the mouth of the active site. These interactions are similarly required for activation of the substrate diphosphate group to initiate catalysis, although partial occupancy binding of the Mg 2+ B ion suggests that this structure represents the penultimate metal coordination complex just prior to substrate activation. The structure of the liganded enzyme enables a precise definition of the enclosed active site contour that serves as a template for the cyclization reaction. This contour is very product-like in shape and readily fits an extended conformation of sesquisabinene and its precursor, the homobisabolyl cation. Structural comparisons of SQS1 with epi-isozizaene synthase mutants that also generate sesquisabinene suggest that [3.1.0] ring formation is not dependent on the isoprenoid tail conformation of the homobisabolyl cation.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry , University of Pennsylvania , 231 South 34th Street , Philadelphia , Pennsylvania 19104-6323 , United States.