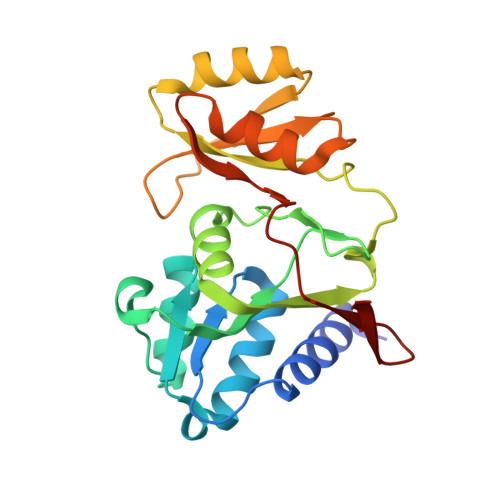
Crystal Structure of Ribose-5-phosphate Isomerase from Legionella pneumophila with Bound Substrate Ribose-5-Phosphate and Product Ribulose-5-Phosphate
Braverman, K.N., Dranow, D.M., Abendroth, J., Lorimer, D.D., Horanyi, P.S., Edwards, T.E.To be published.