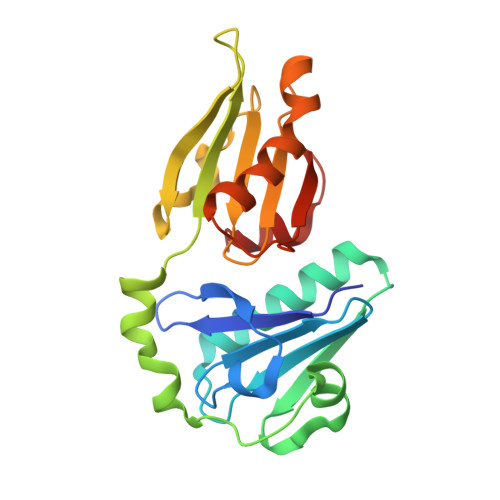
Novel inter-domain Ca2+-binding site in the gelsolin superfamily protein fragmin.
Takeda, S., Fujiwara, I., Sugimoto, Y., Oda, T., Narita, A., Maeda, Y.(2020) J Muscle Res Cell Motil 41: 153-162
- PubMed: 31863323
- DOI: https://doi.org/10.1007/s10974-019-09571-5
- Primary Citation of Related Structures:
6KWZ, 6LJC, 6LJD, 6LJE, 6LJF - PubMed Abstract:
Gelsolin superfamily proteins, consisting of multiple domains (usually six), sever actin filaments and cap the barbed ends in a Ca 2+ -dependent manner. Two types of evolutionally conserved Ca 2+ -binding sites have been identified in this family; type-1 (between gelsolin and actin) and type-2 (within the gelsolin domain). Fragmin, a member in the slime mold Physarum polycephalum, consists of three domains (F1-F3) that are highly similar to the N-terminal half of mammalian gelsolin (G1-G3). Despite their similarities, the two proteins exhibit a significant difference in the Ca 2+ dependency; F1-F3 absolutely requires Ca 2+ for the filament severing whereas G1-G3 does not. In this study, we examined the strong dependency of fragmin on Ca 2+ using biochemical and structural approaches. Our co-sedimentation assay demonstrated that Ca 2+ significantly enhanced the binding of F2-F3 to actin. We determined the crystal structure of F2-F3 in the presence of Ca 2+ . F2-F3 binds a total of three calcium ions; while two are located in type-2 sites within F2 or F3, the remaining one resides between the F2 long helix and the F3 short helix. The inter-domain Ca 2+ -coordination appears to stabilize F2-F3 in a closely packed configuration. Notably, the F3 long helix exhibits a bent conformation which is different from the straight G3 long helix in the presence of Ca 2+ . Our results provide the first structural evidence for the existence of an unconventional Ca 2+ -binding site in the gelsolin superfamily proteins.
Organizational Affiliation:
Structural Biology Research Center, Graduate School of Science, Nagoya University, Furo-cho, Chikusa-ku, Nagoya, 464-8601, Japan. takeda.shuichi@f.mbox.nagoya-u.ac.jp.