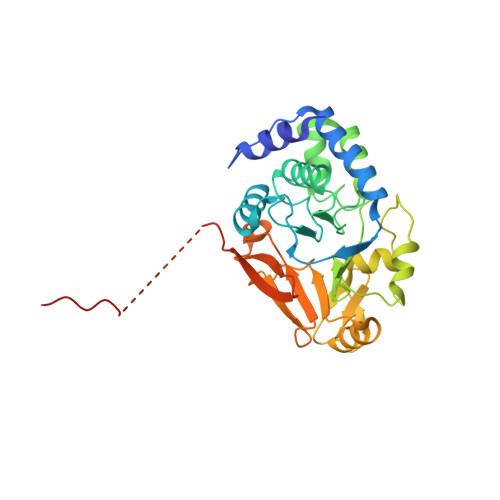
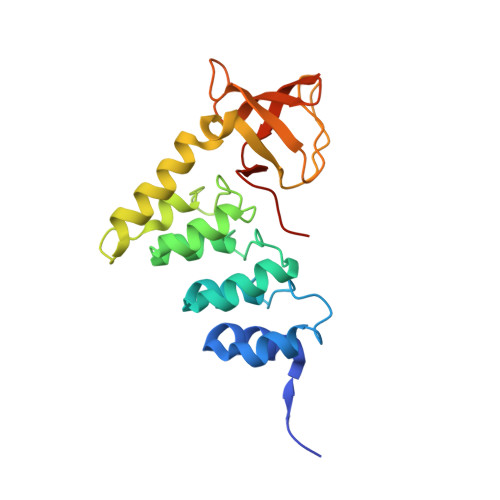
ASPP proteins discriminate between PP1 catalytic subunits through their SH3 domain and the PP1 C-tail.
Bertran, M.T., Mouilleron, S., Zhou, Y., Bajaj, R., Uliana, F., Kumar, G.S., van Drogen, A., Lee, R., Banerjee, J.J., Hauri, S., O'Reilly, N., Gstaiger, M., Page, R., Peti, W., Tapon, N.(2019) Nat Commun 10: 771-771
- PubMed: 30770806
- DOI: https://doi.org/10.1038/s41467-019-08686-0
- Primary Citation of Related Structures:
6GHM - PubMed Abstract:
Serine/threonine phosphatases such as PP1 lack substrate specificity and associate with a large array of targeting subunits to achieve the requisite selectivity. The tumour suppressor ASPP (apoptosis-stimulating protein of p53) proteins associate with PP1 catalytic subunits and are implicated in multiple functions from transcriptional regulation to cell junction remodelling. Here we show that Drosophila ASPP is part of a multiprotein PP1 complex and that PP1 association is necessary for several in vivo functions of Drosophila ASPP. We solve the crystal structure of the human ASPP2/PP1 complex and show that ASPP2 recruits PP1 using both its canonical RVxF motif, which binds the PP1 catalytic domain, and its SH3 domain, which engages the PP1 C-terminal tail. The ASPP2 SH3 domain can discriminate between PP1 isoforms using an acidic specificity pocket in the n-Src domain, providing an exquisite mechanism where multiple motifs are used combinatorially to tune binding affinity to PP1.
Organizational Affiliation:
Apoptosis and Proliferation Control Laboratory, The Francis Crick Institute, 1 Midland Road, London, NW1 1AT, UK.