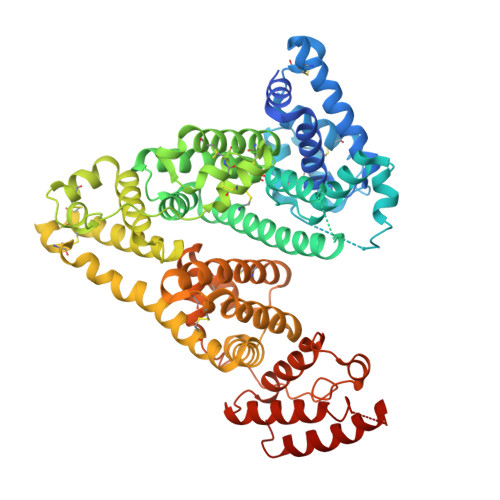
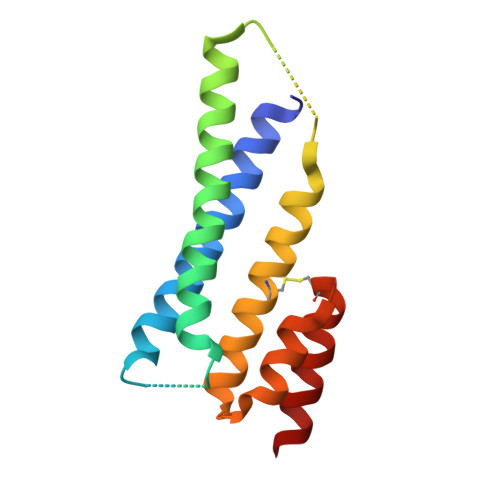
Cell-penetrating Alphabody protein scaffolds for intracellular drug targeting.
Pannecoucke, E., Van Trimpont, M., Desmet, J., Pieters, T., Reunes, L., Demoen, L., Vuylsteke, M., Loverix, S., Vandenbroucke, K., Alard, P., Henderikx, P., Deroo, S., Baatz, F., Lorent, E., Thiolloy, S., Somers, K., McGrath, Y., Van Vlierberghe, P., Lasters, I., Savvides, S.N.(2021) Sci Adv 7
- PubMed: 33771865
- DOI: https://doi.org/10.1126/sciadv.abe1682
- Primary Citation of Related Structures:
6ZIE, 6ZL1 - PubMed Abstract:
The therapeutic scope of antibody and nonantibody protein scaffolds is still prohibitively limited against intracellular drug targets. Here, we demonstrate that the Alphabody scaffold can be engineered into a cell-penetrating protein antagonist against induced myeloid leukemia cell differentiation protein MCL-1, an intracellular target in cancer, by grafting the critical B-cell lymphoma 2 homology 3 helix of MCL-1 onto the Alphabody and tagging the scaffold's termini with designed cell-penetration polypeptides. Introduction of an albumin-binding moiety extended the serum half-life of the engineered Alphabody to therapeutically relevant levels, and administration thereof in mouse tumor xenografts based on myeloma cell lines reduced tumor burden. Crystal structures of such a designed Alphabody in complex with MCL-1 and serum albumin provided the structural blueprint of the applied design principles. Collectively, we provide proof of concept for the use of Alphabodies against intracellular disease mediators, which, to date, have remained in the realm of small-molecule therapeutics.
Organizational Affiliation:
VIB Center for Inflammation Research, 9052 Ghent, Belgium.