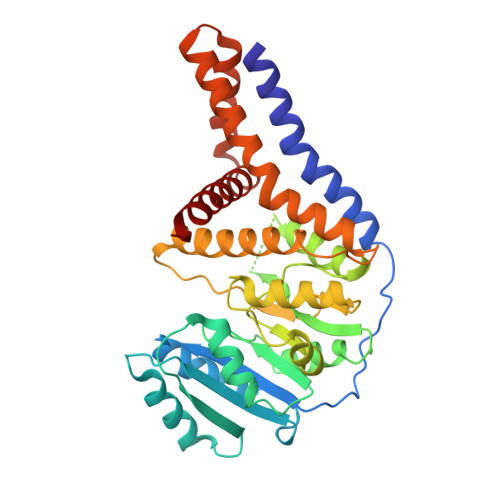
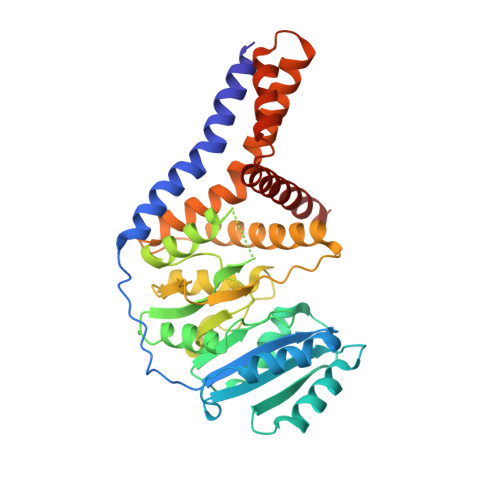
A Transmembrane Crenarchaeal Mannosyltransferase Is Involved in N-Glycan Biosynthesis and Displays an Unexpected Minimal Cellulose-Synthase-like Fold.
Gandini, R., Reichenbach, T., Spadiut, O., Tan, T.C., Kalyani, D.C., Divne, C.(2020) J Mol Biol 432: 4658-4672
- PubMed: 32569746
- DOI: https://doi.org/10.1016/j.jmb.2020.06.016
- Primary Citation of Related Structures:
6YV7, 6YV8, 6YV9 - PubMed Abstract:
Protein glycosylation constitutes a critical post-translational modification that supports a vast number of biological functions in living organisms across all domains of life. A seemingly boundless number of enzymes, glycosyltransferases, are involved in the biosynthesis of these protein-linked glycans. Few glycan-biosynthetic glycosyltransferases have been characterized in vitro, mainly due to the majority being integral membrane proteins and the paucity of relevant acceptor substrates. The crenarchaeote Pyrobaculum calidifontis belongs to the TACK superphylum of archaea (Thaumarchaeota, Aigarchaeota, Crenarchaeota, Korarchaeota) that has been proposed as an eukaryotic ancestor. In archaea, N-glycans are mainly found on cell envelope surface-layer proteins, archaeal flagellins and pili. Archaeal N-glycans are distinct from those of eukaryotes, but one noteworthy exception is the high-mannose N-glycan produced by P. calidifontis, which is similar in sugar composition to the eukaryotic counterpart. Here, we present the characterization and crystal structure of the first member of a crenarchaeal membrane glycosyltransferase, PcManGT. We show that the enzyme is a GDP-, dolichylphosphate-, and manganese-dependent mannosyltransferase. The membrane domain of PcManGT includes three transmembrane helices that topologically coincide with "half" of the six-transmembrane helix cellulose-binding tunnel in Rhodobacter spheroides cellulose synthase BcsA. Conceivably, this "half tunnel" would be suitable for binding the dolichylphosphate-linked acceptor substrate. The PcManGT gene (Pcal_0472) is located in a large gene cluster comprising 14 genes of which 6 genes code for glycosyltransferases, and we hypothesize that this cluster may constitute a crenarchaeal N-glycosylation (PNG) gene cluster.
Organizational Affiliation:
School of Engineering Sciences in Chemistry, Biotechnology, and Health (CBH), KTH Royal Institute of Technology, 10691 Stockholm, Sweden.