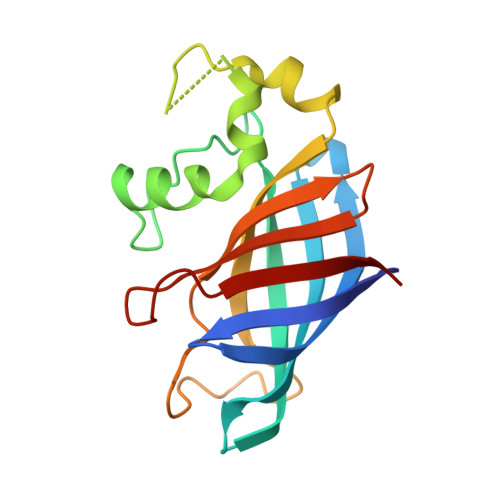
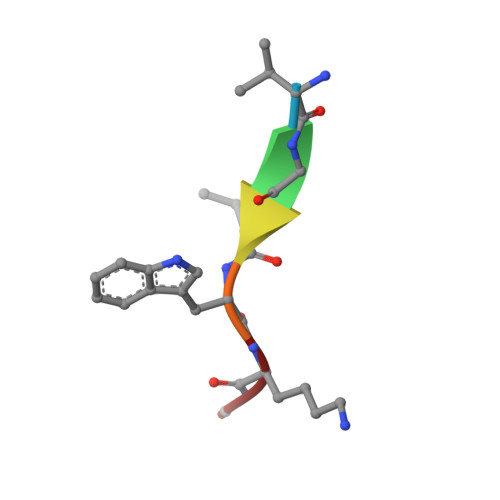
Recognition of nonproline N-terminal residues by the Pro/N-degron pathway.
Dong, C., Chen, S.J., Melnykov, A., Weirich, S., Sun, K., Jeltsch, A., Varshavsky, A., Min, J.(2020) Proc Natl Acad Sci U S A 117: 14158-14167
- PubMed: 32513738
- DOI: https://doi.org/10.1073/pnas.2007085117
- Primary Citation of Related Structures:
6WZX, 6WZZ - PubMed Abstract:
Eukaryotic N-degron pathways are proteolytic systems whose unifying feature is their ability to recognize proteins containing N-terminal (Nt) degradation signals called N-degrons, and to target these proteins for degradation by the 26S proteasome or autophagy. GID4, a subunit of the GID ubiquitin ligase, is the main recognition component of the proline (Pro)/N-degron pathway. GID4 targets proteins through their Nt-Pro residue or a Pro at position 2, in the presence of specific downstream sequence motifs. Here we show that human GID4 can also recognize hydrophobic Nt-residues other than Pro. One example is the sequence Nt-IGLW, bearing Nt-Ile. Nt-IGLW binds to wild-type human GID4 with a K d of 16 μM, whereas the otherwise identical Nt-Pro-bearing sequence PGLW binds to GID4 more tightly, with a K d of 1.9 μM. Despite this difference in affinities of GID4 for Nt-IGLW vs. Nt-PGLW, we found that the GID4-mediated Pro/N-degron pathway of the yeast Saccharomyces cerevisiae can target an Nt-IGLW-bearing protein for rapid degradation. We solved crystal structures of human GID4 bound to a peptide bearing Nt-Ile or Nt-Val. We also altered specific residues of human GID4 and measured the affinities of resulting mutant GID4s for Nt-IGLW and Nt-PGLW, thereby determining relative contributions of specific GID4 residues to the GID4-mediated recognition of Nt-Pro vs. Nt-residues other than Pro. These and related results advance the understanding of targeting by the Pro/N-degron pathway and greatly expand the substrate recognition range of the GID ubiquitin ligase in both human and yeast cells.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, 300070 Tianjin, People's Republic of China.